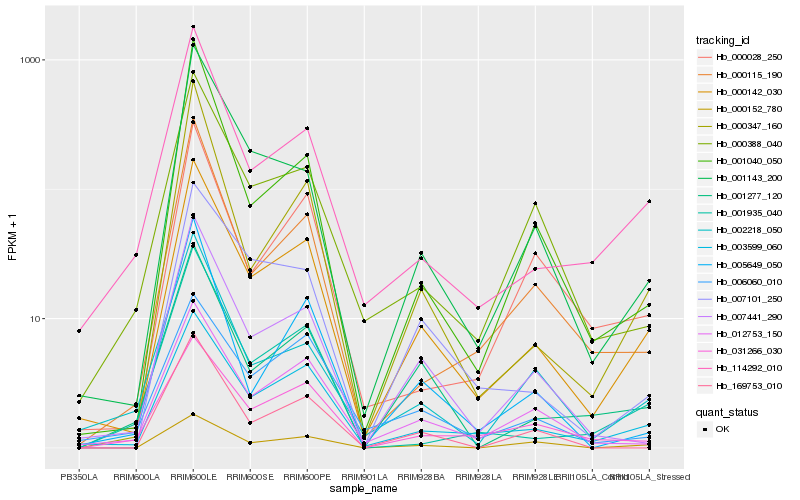
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000142_030 |
0.0 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_003599_060 |
0.0822684419 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 3 |
Hb_114292_010 |
0.1270341287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007441_290 |
0.1276263809 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 5 |
Hb_012753_150 |
0.1320190277 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 6 |
Hb_001277_120 |
0.1388698414 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 7 |
Hb_001143_200 |
0.1392692573 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 8 |
Hb_007101_250 |
0.1471032601 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 9 |
Hb_000115_190 |
0.1488824919 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 10 |
Hb_000347_160 |
0.148946768 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 11 |
Hb_000388_040 |
0.1504120991 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 12 |
Hb_001935_040 |
0.1511603007 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 8 [Jatropha curcas] |
| 13 |
Hb_002218_050 |
0.1520177843 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 14 |
Hb_000152_780 |
0.1535534809 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 15 |
Hb_169753_010 |
0.1548826444 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 16 |
Hb_031266_030 |
0.1578311653 |
- |
- |
Kinase superfamily protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_001040_050 |
0.1593148725 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 18 |
Hb_005649_050 |
0.1593253225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000028_250 |
0.1598377642 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 20 |
Hb_006060_010 |
0.1612099996 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |