| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
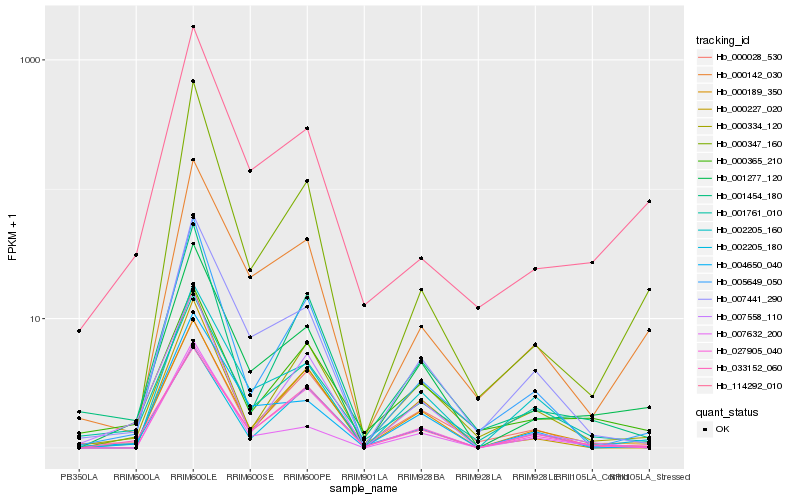
Hb_001277_120 |
0.0 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 2 |
Hb_000189_350 |
0.1331946295 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 3 [Jatropha curcas] |
| 3 |
Hb_000142_030 |
0.1388698414 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_007441_290 |
0.1453150049 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 5 |
Hb_004650_040 |
0.1479729185 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 6 |
Hb_114292_010 |
0.1563616944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001761_010 |
0.1566276837 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 8 |
Hb_033152_060 |
0.1584331983 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 [Jatropha curcas] |
| 9 |
Hb_000347_160 |
0.1656896702 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 10 |
Hb_002205_180 |
0.168169217 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 11 |
Hb_007558_110 |
0.1688865892 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 12 |
Hb_000365_210 |
0.169009279 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 13 |
Hb_027905_040 |
0.1710167972 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: E2F transcription factor-like E2FF [Jatropha curcas] |
| 14 |
Hb_005649_050 |
0.1720611721 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000028_530 |
0.1723259357 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 16 |
Hb_001454_180 |
0.173370374 |
- |
- |
PREDICTED: uncharacterized protein LOC105643432 [Jatropha curcas] |
| 17 |
Hb_002205_160 |
0.17513868 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 18 |
Hb_007632_200 |
0.1762969193 |
- |
- |
hypothetical protein L484_018221 [Morus notabilis] |
| 19 |
Hb_000334_120 |
0.17659333 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 20 |
Hb_000227_020 |
0.1773947818 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |