| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
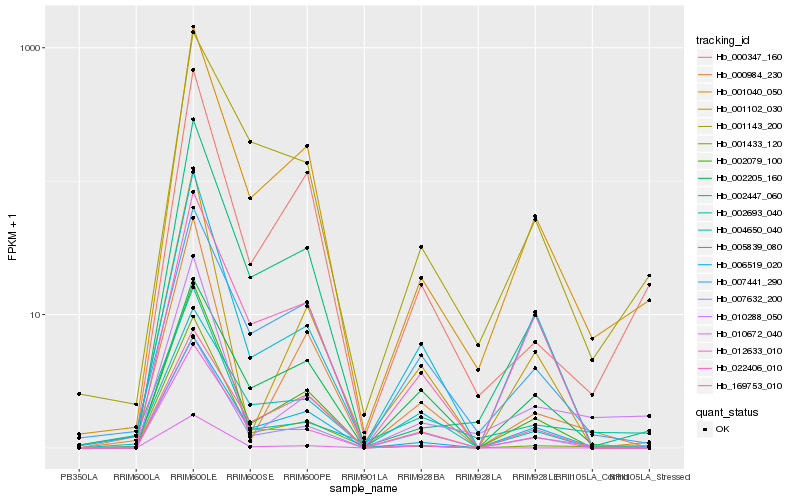
Hb_007632_200 |
0.0 |
- |
- |
hypothetical protein L484_018221 [Morus notabilis] |
| 2 |
Hb_001040_050 |
0.112720323 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 3 |
Hb_005839_080 |
0.1154160221 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 4 |
Hb_004650_040 |
0.135447558 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 5 |
Hb_012633_010 |
0.1379796483 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 6 |
Hb_006519_020 |
0.1399928115 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 7 |
Hb_001102_030 |
0.1437825631 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 8 |
Hb_007441_290 |
0.1468430775 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 9 |
Hb_010672_040 |
0.147952157 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 10 |
Hb_000347_160 |
0.1520834525 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 11 |
Hb_000984_230 |
0.15386378 |
- |
- |
- |
| 12 |
Hb_002079_100 |
0.1547637147 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 13 |
Hb_002447_060 |
0.1585020987 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_022406_010 |
0.1589516551 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_010288_050 |
0.1592700477 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 16 |
Hb_001433_120 |
0.159290549 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 17 |
Hb_001143_200 |
0.1608018242 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 18 |
Hb_002693_040 |
0.1613368647 |
- |
- |
hypersensitive-induced response protein [Carica papaya] |
| 19 |
Hb_169753_010 |
0.1624579263 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 20 |
Hb_002205_160 |
0.1636904717 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |