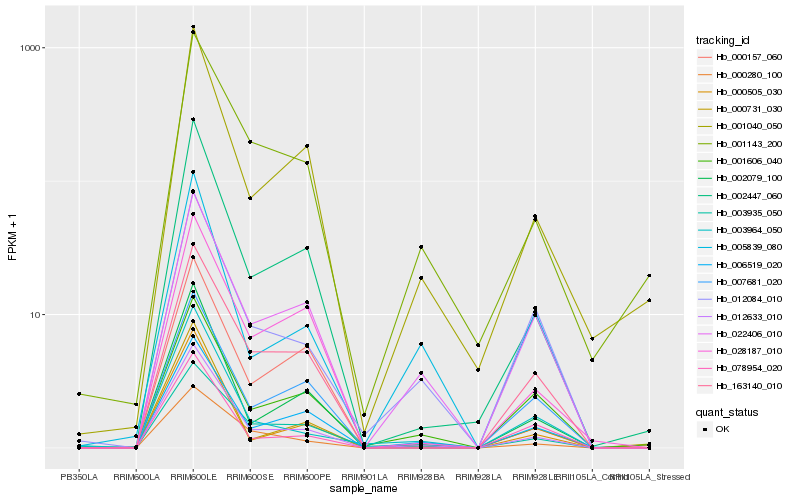
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012633_010 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 2 |
Hb_002447_060 |
0.0732886847 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002079_100 |
0.0889367599 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 4 |
Hb_006519_020 |
0.1028817059 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 5 |
Hb_001040_050 |
0.1068863986 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 6 |
Hb_000505_030 |
0.1142409207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_163140_010 |
0.1147850802 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 8 |
Hb_000280_100 |
0.1192235184 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |
| 9 |
Hb_003964_050 |
0.1222621375 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 25-like [Jatropha curcas] |
| 10 |
Hb_022406_010 |
0.1263993342 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_028187_010 |
0.1272141291 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 12 |
Hb_012084_010 |
0.1301168945 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 13 |
Hb_007681_020 |
0.1315521362 |
- |
- |
transferase, putative [Ricinus communis] |
| 14 |
Hb_000731_030 |
0.1331559531 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_005839_080 |
0.1351177898 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 16 |
Hb_003935_050 |
0.1351782809 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_001143_200 |
0.1353773038 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 18 |
Hb_000157_060 |
0.1354244197 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001606_040 |
0.136662987 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 20 |
Hb_078954_020 |
0.1371388883 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |