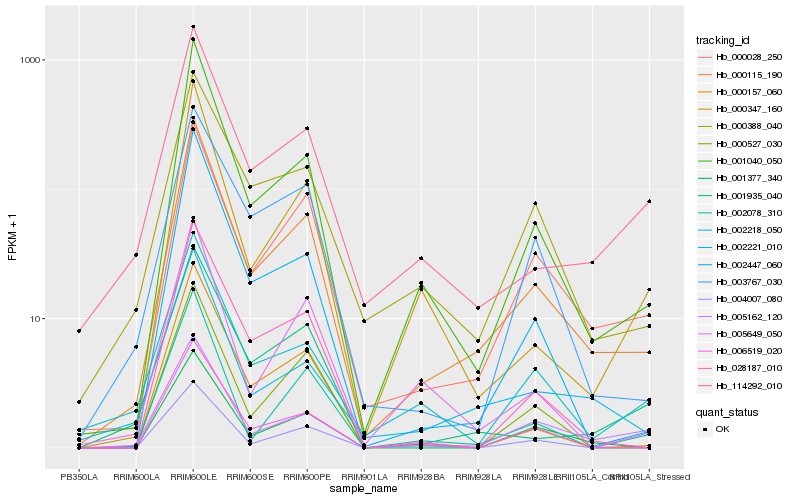
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000115_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 2 |
Hb_001040_050 |
0.0870484705 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 3 |
Hb_000028_250 |
0.1045142898 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 4 |
Hb_006519_020 |
0.1296946322 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 5 |
Hb_002447_060 |
0.1306674178 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000388_040 |
0.1310895481 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 7 |
Hb_005162_120 |
0.1311902691 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 8 |
Hb_002221_010 |
0.1328872297 |
- |
- |
PREDICTED: adenylyl-sulfate kinase 3-like [Jatropha curcas] |
| 9 |
Hb_028187_010 |
0.1353087225 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 10 |
Hb_001377_340 |
0.135829552 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF023 [Jatropha curcas] |
| 11 |
Hb_114292_010 |
0.1372626736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002078_310 |
0.1399643367 |
- |
- |
PREDICTED: uncharacterized protein LOC105645005 [Jatropha curcas] |
| 13 |
Hb_000157_060 |
0.1408102349 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000347_160 |
0.1418314607 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 15 |
Hb_005649_050 |
0.1422068761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004007_080 |
0.1444064335 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g49900 [Jatropha curcas] |
| 17 |
Hb_001935_040 |
0.1447008652 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 8 [Jatropha curcas] |
| 18 |
Hb_000527_030 |
0.1448337944 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 19 |
Hb_003767_030 |
0.1464230416 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 20 |
Hb_002218_050 |
0.147484769 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |