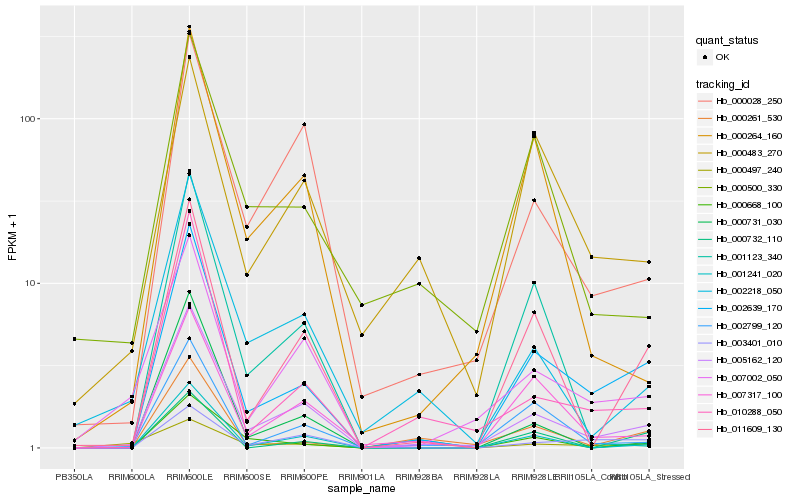
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002639_170 |
0.0 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 2 |
Hb_005162_120 |
0.1397556711 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 3 |
Hb_011609_130 |
0.1684288512 |
- |
- |
- |
| 4 |
Hb_000261_530 |
0.1746347053 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 5 |
Hb_010288_050 |
0.1921771206 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 6 |
Hb_001241_020 |
0.1940955778 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 7 |
Hb_007317_100 |
0.1942887683 |
- |
- |
Peflin, putative [Ricinus communis] |
| 8 |
Hb_000668_100 |
0.1994218521 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 9 |
Hb_003401_010 |
0.2003875155 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 10 |
Hb_002799_120 |
0.2017240524 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 11 |
Hb_000732_110 |
0.2040426097 |
- |
- |
hypothetical protein JCGZ_21339 [Jatropha curcas] |
| 12 |
Hb_007002_050 |
0.2271405863 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 13 |
Hb_000483_270 |
0.2315625345 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 14 |
Hb_000264_160 |
0.2352459198 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 15 |
Hb_002218_050 |
0.2371349846 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 16 |
Hb_000731_030 |
0.2377578257 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000497_240 |
0.2391235705 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 18 |
Hb_000028_250 |
0.2410572228 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 19 |
Hb_000500_330 |
0.2424908494 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 20 |
Hb_001123_340 |
0.244016035 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |