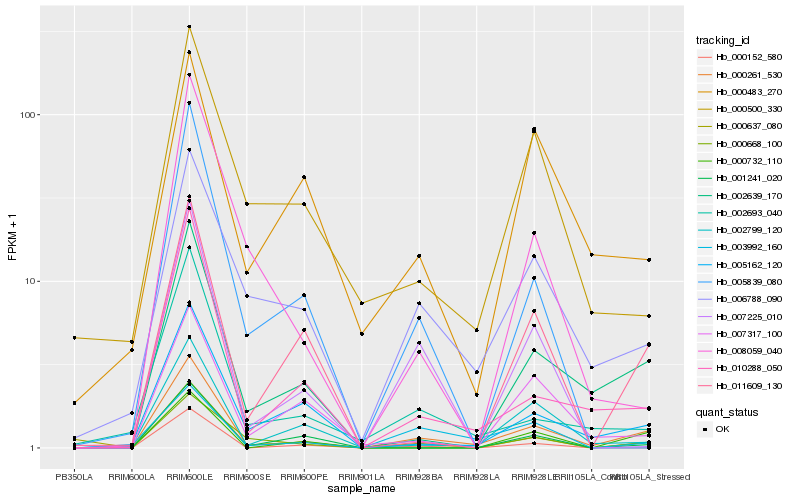
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_530 |
0.0 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 2 |
Hb_002639_170 |
0.1746347053 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 3 |
Hb_010288_050 |
0.1905827106 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 4 |
Hb_006788_090 |
0.2065109833 |
- |
- |
- |
| 5 |
Hb_005162_120 |
0.2073864982 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 6 |
Hb_011609_130 |
0.2111605359 |
- |
- |
- |
| 7 |
Hb_002799_120 |
0.2149927521 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 8 |
Hb_002693_040 |
0.2177490029 |
- |
- |
hypersensitive-induced response protein [Carica papaya] |
| 9 |
Hb_000637_080 |
0.2231597087 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 10 |
Hb_000732_110 |
0.225370487 |
- |
- |
hypothetical protein JCGZ_21339 [Jatropha curcas] |
| 11 |
Hb_007225_010 |
0.2314959979 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 12 |
Hb_007317_100 |
0.2404064163 |
- |
- |
Peflin, putative [Ricinus communis] |
| 13 |
Hb_001241_020 |
0.2413704056 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 14 |
Hb_000500_330 |
0.2422568188 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 15 |
Hb_000668_100 |
0.2424140447 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 16 |
Hb_005839_080 |
0.2462969404 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 17 |
Hb_003992_160 |
0.2463382491 |
- |
- |
PREDICTED: cucumber peeling cupredoxin-like [Jatropha curcas] |
| 18 |
Hb_008059_040 |
0.2490484 |
- |
- |
hypothetical protein POPTR_0019s12360g [Populus trichocarpa] |
| 19 |
Hb_000483_270 |
0.2504946799 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 20 |
Hb_000152_580 |
0.2514748142 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |