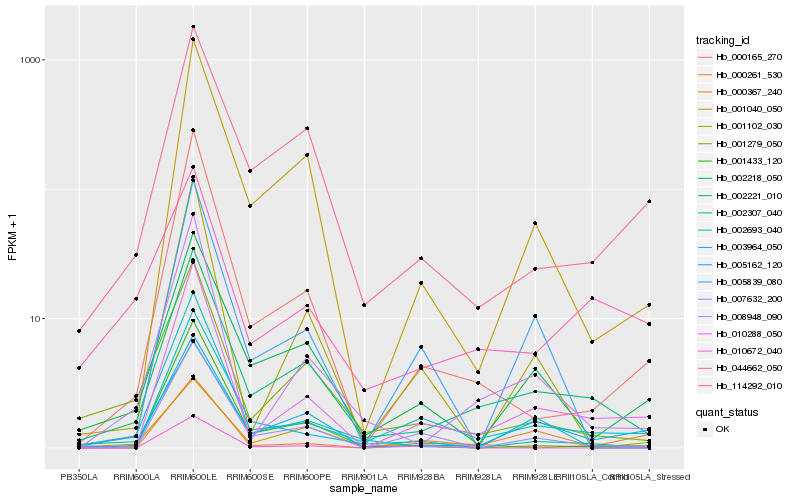
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002693_040 |
0.0 |
- |
- |
hypersensitive-induced response protein [Carica papaya] |
| 2 |
Hb_010288_050 |
0.1347496488 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 3 |
Hb_002221_010 |
0.1599522295 |
- |
- |
PREDICTED: adenylyl-sulfate kinase 3-like [Jatropha curcas] |
| 4 |
Hb_007632_200 |
0.1613368647 |
- |
- |
hypothetical protein L484_018221 [Morus notabilis] |
| 5 |
Hb_000165_270 |
0.1643863934 |
transcription factor |
TF Family: Tify |
JAZ10 [Hevea brasiliensis] |
| 6 |
Hb_002218_050 |
0.175460762 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 7 |
Hb_008948_090 |
0.1828360998 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_001279_050 |
0.1840802246 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 9 |
Hb_005839_080 |
0.190522676 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 10 |
Hb_114292_010 |
0.2001171493 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001040_050 |
0.2009331298 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 12 |
Hb_001102_030 |
0.2024830273 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 13 |
Hb_001433_120 |
0.2035808627 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 14 |
Hb_044662_050 |
0.2042464696 |
- |
- |
hypothetical protein POPTR_0002s08440g [Populus trichocarpa] |
| 15 |
Hb_005162_120 |
0.2081141182 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 16 |
Hb_003964_050 |
0.20871321 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 25-like [Jatropha curcas] |
| 17 |
Hb_002307_040 |
0.2104850868 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000367_240 |
0.2126929368 |
- |
- |
hypothetical protein JCGZ_11682 [Jatropha curcas] |
| 19 |
Hb_000261_530 |
0.2177490029 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 20 |
Hb_010672_040 |
0.2180217582 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |