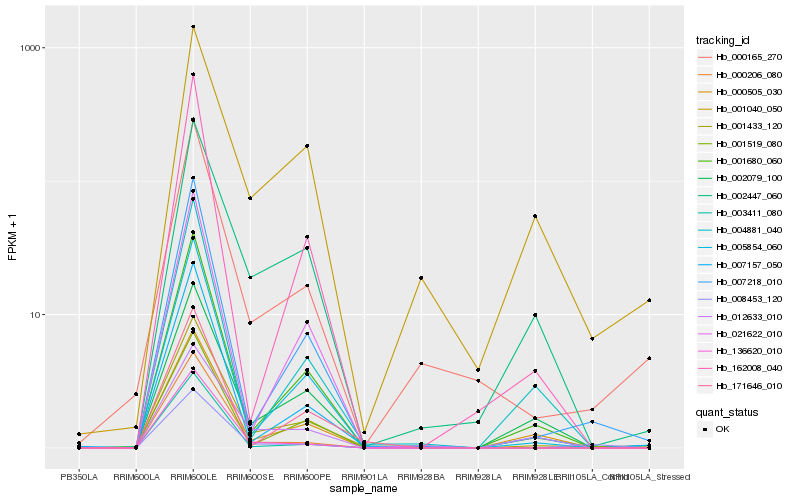
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001433_120 |
0.0 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 2 |
Hb_000165_270 |
0.093563032 |
transcription factor |
TF Family: Tify |
JAZ10 [Hevea brasiliensis] |
| 3 |
Hb_007218_010 |
0.1158361518 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X1 [Populus euphratica] |
| 4 |
Hb_005854_060 |
0.1179765827 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_007157_050 |
0.1182727189 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 6 |
Hb_001680_060 |
0.1204545189 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002447_060 |
0.1262612498 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_136620_010 |
0.1285558028 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 9 |
Hb_000206_080 |
0.1314703728 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 10 |
Hb_001040_050 |
0.1346025147 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 11 |
Hb_001519_080 |
0.1347722293 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 12 |
Hb_002079_100 |
0.135527851 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 13 |
Hb_000505_030 |
0.1359545576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_021622_010 |
0.1412406444 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |
| 15 |
Hb_012633_010 |
0.1431990811 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 16 |
Hb_171646_010 |
0.143418694 |
- |
- |
hypothetical protein B456_012G144300, partial [Gossypium raimondii] |
| 17 |
Hb_162008_040 |
0.14543673 |
- |
- |
hypothetical protein CL1725Contig1_03, partial [Pinus taeda] |
| 18 |
Hb_003411_080 |
0.1461046074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_008453_120 |
0.1463682973 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0005s26480g [Populus trichocarpa] |
| 20 |
Hb_004881_040 |
0.1463903663 |
- |
- |
PREDICTED: protodermal factor 1-like [Populus euphratica] |