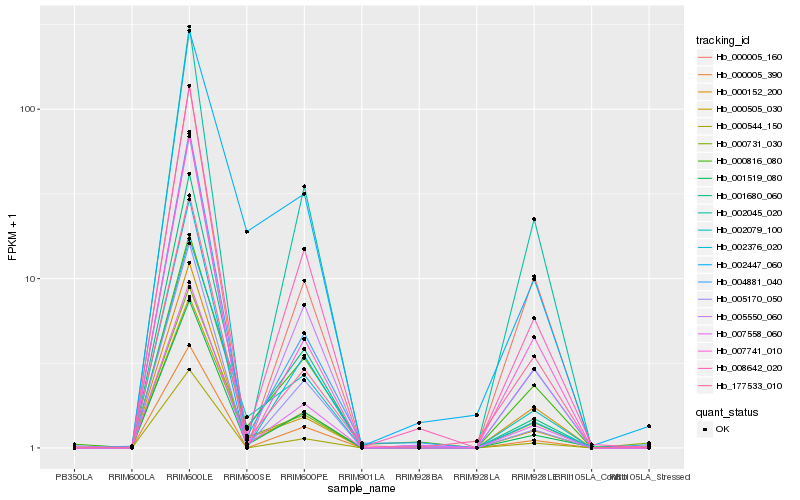
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000505_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002079_100 |
0.0535666808 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 3 |
Hb_000731_030 |
0.0666317032 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_007558_060 |
0.0768664722 |
- |
- |
PREDICTED: uncharacterized protein LOC105644482 [Jatropha curcas] |
| 5 |
Hb_000152_200 |
0.0808647303 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase ERL2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001680_060 |
0.0822192395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001519_080 |
0.0829530168 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 8 |
Hb_004881_040 |
0.0931206437 |
- |
- |
PREDICTED: protodermal factor 1-like [Populus euphratica] |
| 9 |
Hb_008642_020 |
0.0942088162 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 10 |
Hb_000544_150 |
0.0960788176 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 1 [Jatropha curcas] |
| 11 |
Hb_005550_060 |
0.0996163366 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_002447_060 |
0.0997941007 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000005_390 |
0.1046567085 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 14 |
Hb_007741_010 |
0.1078332461 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 1 [Jatropha curcas] |
| 15 |
Hb_177533_010 |
0.10867848 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 16 |
Hb_000005_160 |
0.1096948431 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 17 |
Hb_005170_050 |
0.1104289146 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Populus euphratica] |
| 18 |
Hb_000816_080 |
0.1133734182 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 19 |
Hb_002045_020 |
0.1134863161 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 20 |
Hb_002376_020 |
0.1138274134 |
- |
- |
lectin, partial [Lathyrus palustris] |