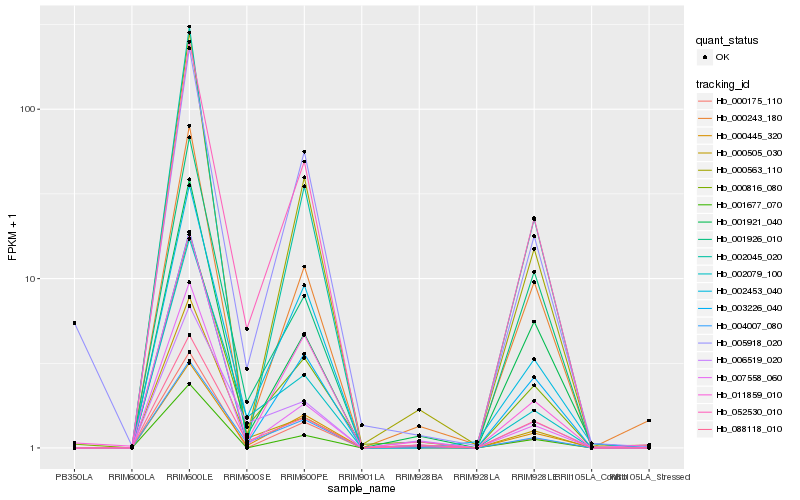
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_080 |
0.0 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 2 |
Hb_088118_010 |
0.0754043208 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 3 |
Hb_052530_010 |
0.080466466 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002079_100 |
0.0898474559 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 5 |
Hb_003226_040 |
0.0911209718 |
- |
- |
PREDICTED: endoglucanase 17 [Populus euphratica] |
| 6 |
Hb_004007_080 |
0.0927033086 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g49900 [Jatropha curcas] |
| 7 |
Hb_007558_060 |
0.0980586475 |
- |
- |
PREDICTED: uncharacterized protein LOC105644482 [Jatropha curcas] |
| 8 |
Hb_011859_010 |
0.1028179373 |
- |
- |
hypothetical protein VITISV_027923 [Vitis vinifera] |
| 9 |
Hb_000243_180 |
0.1029050721 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1 [Jatropha curcas] |
| 10 |
Hb_001921_040 |
0.1029842404 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 11 |
Hb_000563_110 |
0.1047090701 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 12 |
Hb_000445_320 |
0.1099853146 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |
| 13 |
Hb_001926_010 |
0.1104401688 |
- |
- |
hypothetical protein Csa_2G383910 [Cucumis sativus] |
| 14 |
Hb_002045_020 |
0.1123026299 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 15 |
Hb_002453_040 |
0.1123922661 |
- |
- |
PREDICTED: uncharacterized protein LOC105636487 [Jatropha curcas] |
| 16 |
Hb_000175_110 |
0.1127209353 |
- |
- |
hypothetical protein JCGZ_06125 [Jatropha curcas] |
| 17 |
Hb_001677_070 |
0.1131535201 |
- |
- |
PREDICTED: putative caffeoyl-CoA O-methyltransferase At1g67980 [Jatropha curcas] |
| 18 |
Hb_000505_030 |
0.1133734182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005918_020 |
0.113819999 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 20 |
Hb_006519_020 |
0.1147056301 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |