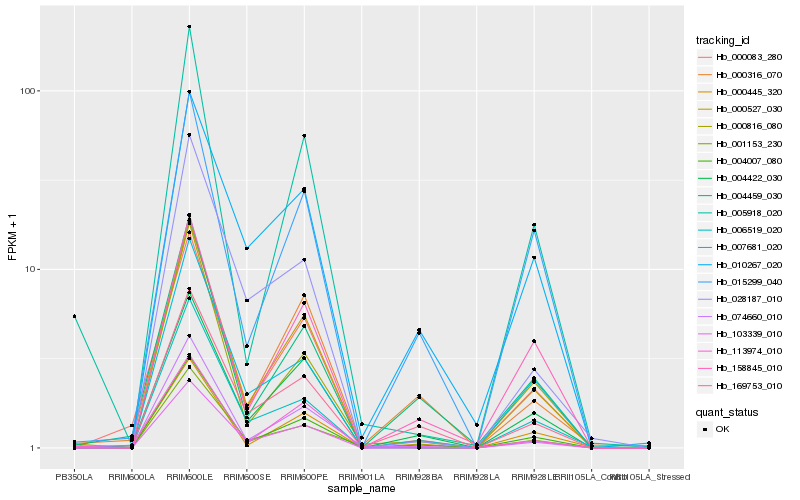
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_103339_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_004007_080 |
0.0952802519 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g49900 [Jatropha curcas] |
| 3 |
Hb_000527_030 |
0.0955474439 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 4 |
Hb_004422_030 |
0.1076149802 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK7-like [Jatropha curcas] |
| 5 |
Hb_006519_020 |
0.1134652083 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 6 |
Hb_169753_010 |
0.1145559525 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 7 |
Hb_000083_280 |
0.1261250668 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_005918_020 |
0.1263415073 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 9 |
Hb_010267_020 |
0.1298489622 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 10 |
Hb_000816_080 |
0.1313279845 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 11 |
Hb_158845_010 |
0.1322963808 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 [Jatropha curcas] |
| 12 |
Hb_074660_010 |
0.1331304482 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 13 |
Hb_113974_010 |
0.1331326106 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_028187_010 |
0.1338896287 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 15 |
Hb_007681_020 |
0.1346725066 |
- |
- |
transferase, putative [Ricinus communis] |
| 16 |
Hb_004459_030 |
0.1356701293 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001153_230 |
0.1365157128 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 18 |
Hb_000445_320 |
0.1368061296 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |
| 19 |
Hb_000316_070 |
0.1374606497 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 20 |
Hb_015299_040 |
0.1410335912 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |