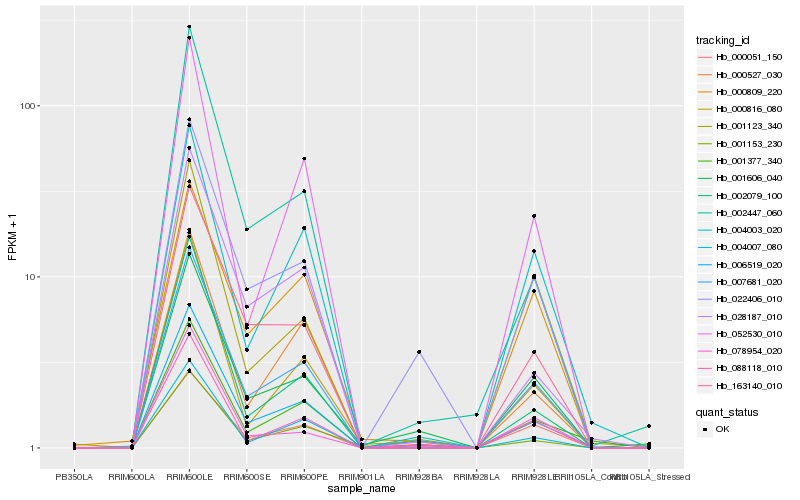
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007681_020 |
0.0 |
- |
- |
transferase, putative [Ricinus communis] |
| 2 |
Hb_163140_010 |
0.0773634733 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 3 |
Hb_000051_150 |
0.0895714507 |
- |
- |
hypothetical protein CICLE_v10003316mg [Citrus clementina] |
| 4 |
Hb_006519_020 |
0.0935470707 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 5 |
Hb_001377_340 |
0.0983045896 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF023 [Jatropha curcas] |
| 6 |
Hb_001153_230 |
0.1016794467 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 7 |
Hb_088118_010 |
0.1061027028 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 8 |
Hb_052530_010 |
0.1079626261 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001606_040 |
0.1084984743 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 10 |
Hb_000527_030 |
0.1106116623 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 11 |
Hb_022406_010 |
0.1134958735 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_002079_100 |
0.1157389319 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 13 |
Hb_000816_080 |
0.1163626743 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 14 |
Hb_004003_020 |
0.1174630828 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_028187_010 |
0.1189274344 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 16 |
Hb_004007_080 |
0.120326534 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g49900 [Jatropha curcas] |
| 17 |
Hb_002447_060 |
0.1207756995 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_078954_020 |
0.1243664208 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 19 |
Hb_000809_220 |
0.1244861907 |
- |
- |
PREDICTED: 3-ketodihydrosphingosine reductase [Jatropha curcas] |
| 20 |
Hb_001123_340 |
0.126342941 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |