| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
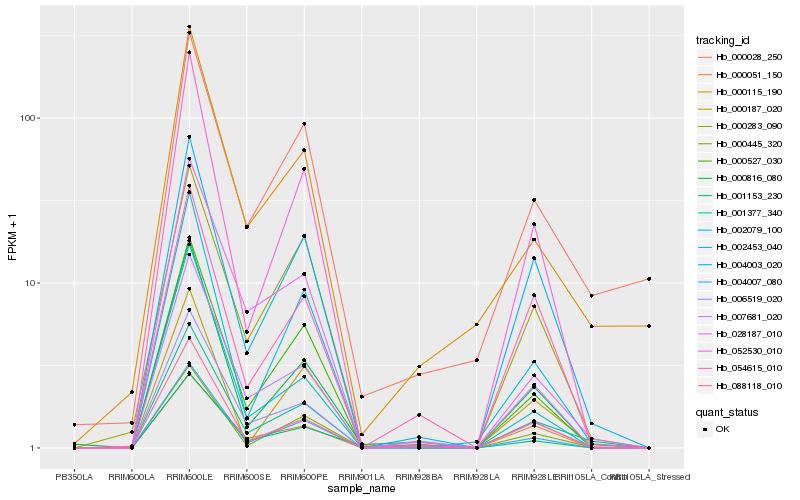
Hb_001377_340 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF023 [Jatropha curcas] |
| 2 |
Hb_007681_020 |
0.0983045896 |
- |
- |
transferase, putative [Ricinus communis] |
| 3 |
Hb_004003_020 |
0.1013778986 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_052530_010 |
0.1136557 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 isoform X2 [Jatropha curcas] |
| 5 |
Hb_006519_020 |
0.1170113929 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 6 |
Hb_000445_320 |
0.1173488044 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |
| 7 |
Hb_000527_030 |
0.1189514156 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 8 |
Hb_002453_040 |
0.1210088429 |
- |
- |
PREDICTED: uncharacterized protein LOC105636487 [Jatropha curcas] |
| 9 |
Hb_001153_230 |
0.1288399763 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 10 |
Hb_088118_010 |
0.1296264426 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 11 |
Hb_000051_150 |
0.1308285851 |
- |
- |
hypothetical protein CICLE_v10003316mg [Citrus clementina] |
| 12 |
Hb_004007_080 |
0.1309000914 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g49900 [Jatropha curcas] |
| 13 |
Hb_000816_080 |
0.1324817956 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 14 |
Hb_000283_090 |
0.1347866333 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000115_190 |
0.135829552 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 16 |
Hb_028187_010 |
0.1403063377 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 17 |
Hb_054615_010 |
0.1412064555 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 18 |
Hb_000187_020 |
0.1414799584 |
- |
- |
Xanthine/uracil permease family protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_000028_250 |
0.1419181769 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 20 |
Hb_002079_100 |
0.1432242905 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |