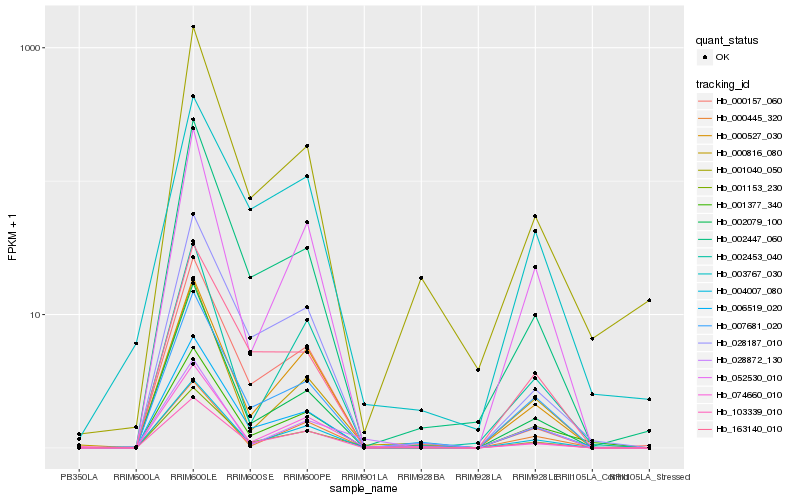
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001153_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 2 |
Hb_074660_010 |
0.0966737962 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 3 |
Hb_000527_030 |
0.1015281825 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 4 |
Hb_007681_020 |
0.1016794467 |
- |
- |
transferase, putative [Ricinus communis] |
| 5 |
Hb_028187_010 |
0.1136522108 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 6 |
Hb_000816_080 |
0.1155791969 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 7 |
Hb_052530_010 |
0.1193754571 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002079_100 |
0.1198226421 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 9 |
Hb_004007_080 |
0.1208734619 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g49900 [Jatropha curcas] |
| 10 |
Hb_006519_020 |
0.1228393965 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 11 |
Hb_002447_060 |
0.1228984362 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000157_060 |
0.1229487858 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002453_040 |
0.1286380327 |
- |
- |
PREDICTED: uncharacterized protein LOC105636487 [Jatropha curcas] |
| 14 |
Hb_001377_340 |
0.1288399763 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF023 [Jatropha curcas] |
| 15 |
Hb_163140_010 |
0.1325663959 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 16 |
Hb_103339_010 |
0.1365157128 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_001040_050 |
0.1405183297 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 18 |
Hb_000445_320 |
0.1418772286 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |
| 19 |
Hb_028872_130 |
0.1419419358 |
- |
- |
PREDICTED: uncharacterized protein LOC105644764 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003767_030 |
0.1455252779 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |