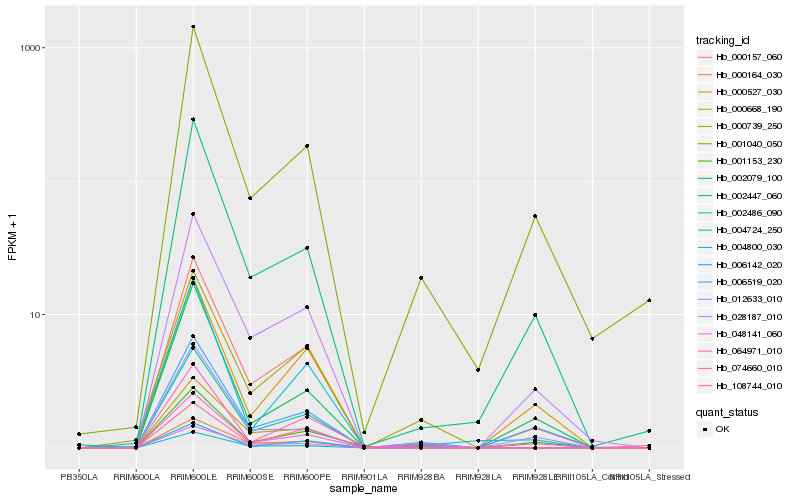
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000157_060 |
0.0 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_028187_010 |
0.062487451 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 3 |
Hb_064971_010 |
0.0915922936 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 4 |
Hb_002447_060 |
0.092288013 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_074660_010 |
0.1021593842 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 6 |
Hb_000164_030 |
0.1049588849 |
- |
- |
PREDICTED: RING-H2 finger protein ATL14-like [Zea mays] |
| 7 |
Hb_006142_020 |
0.1071745657 |
- |
- |
JHL25P11.12 [Jatropha curcas] |
| 8 |
Hb_002486_090 |
0.11341453 |
- |
- |
hypothetical protein POPTR_0005s06910g [Populus trichocarpa] |
| 9 |
Hb_001040_050 |
0.1144733255 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 10 |
Hb_004724_250 |
0.1145321604 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 11 |
Hb_000527_030 |
0.1196118492 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 12 |
Hb_048141_060 |
0.1208701426 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 13 |
Hb_002079_100 |
0.1226672427 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 14 |
Hb_001153_230 |
0.1229487858 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 15 |
Hb_004800_030 |
0.1270067877 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 16 |
Hb_006519_020 |
0.1314071521 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 17 |
Hb_000668_190 |
0.1322367544 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 18 |
Hb_108744_010 |
0.1337034314 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 19 |
Hb_012633_010 |
0.1354244197 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 20 |
Hb_000739_250 |
0.1355922623 |
- |
- |
PREDICTED: uncharacterized protein LOC105120821 [Populus euphratica] |