| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
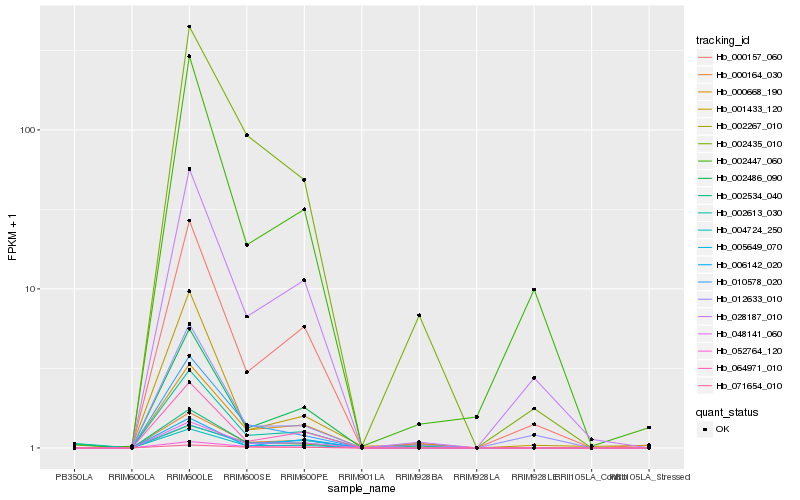
Hb_004724_250 |
0.0 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 2 |
Hb_000164_030 |
0.0551528146 |
- |
- |
PREDICTED: RING-H2 finger protein ATL14-like [Zea mays] |
| 3 |
Hb_048141_060 |
0.0559533934 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 4 |
Hb_010578_020 |
0.0740626401 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_064971_010 |
0.0743763366 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 6 |
Hb_002267_010 |
0.0972207643 |
- |
- |
hypothetical protein SORBIDRAFT_01g040370 [Sorghum bicolor] |
| 7 |
Hb_052764_120 |
0.1003360149 |
- |
- |
PREDICTED: uncharacterized protein LOC103941142 [Pyrus x bretschneideri] |
| 8 |
Hb_000157_060 |
0.1145321604 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000668_190 |
0.1147339107 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 10 |
Hb_002613_030 |
0.1147455696 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002486_090 |
0.1153389706 |
- |
- |
hypothetical protein POPTR_0005s06910g [Populus trichocarpa] |
| 12 |
Hb_002435_010 |
0.1179839451 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 13 |
Hb_002534_040 |
0.1258250275 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 14 |
Hb_028187_010 |
0.1311061741 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 15 |
Hb_006142_020 |
0.1344171354 |
- |
- |
JHL25P11.12 [Jatropha curcas] |
| 16 |
Hb_002447_060 |
0.1357451814 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005649_070 |
0.1367113212 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_071654_010 |
0.1402651705 |
- |
- |
non-ltr retroelement reverse transcriptase [Rosa rugosa] |
| 19 |
Hb_001433_120 |
0.1553093396 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 20 |
Hb_012633_010 |
0.1582867741 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |