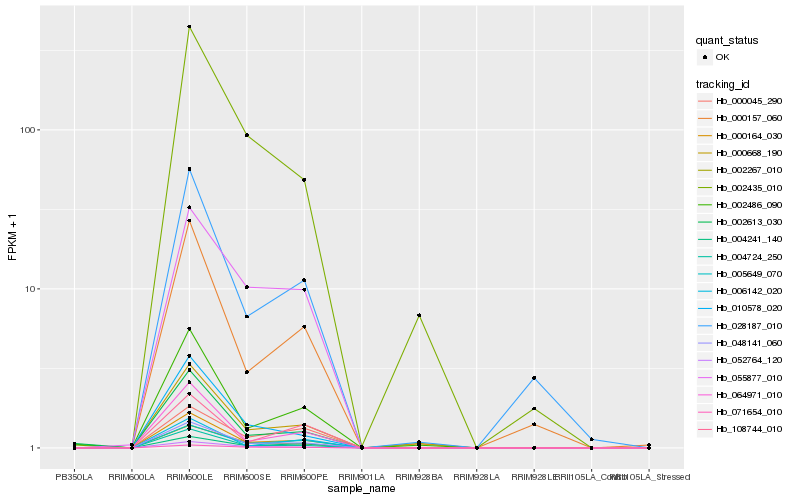
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000164_030 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL14-like [Zea mays] |
| 2 |
Hb_048141_060 |
0.0270960458 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 3 |
Hb_004724_250 |
0.0551528146 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 4 |
Hb_064971_010 |
0.0766284757 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 5 |
Hb_005649_070 |
0.082121642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002267_010 |
0.0822148327 |
- |
- |
hypothetical protein SORBIDRAFT_01g040370 [Sorghum bicolor] |
| 7 |
Hb_000157_060 |
0.1049588849 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000668_190 |
0.105037925 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 9 |
Hb_071654_010 |
0.1064571181 |
- |
- |
non-ltr retroelement reverse transcriptase [Rosa rugosa] |
| 10 |
Hb_052764_120 |
0.1096205088 |
- |
- |
PREDICTED: uncharacterized protein LOC103941142 [Pyrus x bretschneideri] |
| 11 |
Hb_004241_140 |
0.1105309848 |
- |
- |
S-locus lectin protein kinase family protein, putative [Theobroma cacao] |
| 12 |
Hb_002486_090 |
0.11158559 |
- |
- |
hypothetical protein POPTR_0005s06910g [Populus trichocarpa] |
| 13 |
Hb_006142_020 |
0.1119072883 |
- |
- |
JHL25P11.12 [Jatropha curcas] |
| 14 |
Hb_010578_020 |
0.1157594357 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_028187_010 |
0.1193359396 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 16 |
Hb_002435_010 |
0.1254562222 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 17 |
Hb_002613_030 |
0.1254922915 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_108744_010 |
0.1294755396 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 19 |
Hb_055877_010 |
0.131451675 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 20 |
Hb_000045_290 |
0.1350483974 |
- |
- |
- |