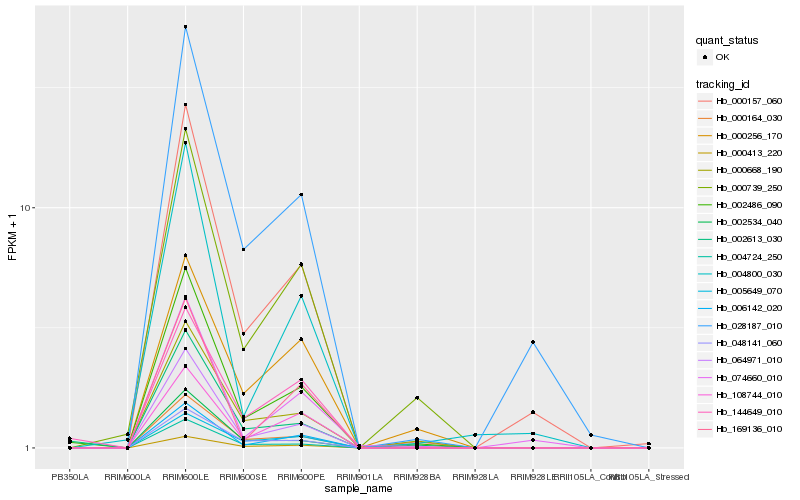
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002486_090 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s06910g [Populus trichocarpa] |
| 2 |
Hb_064971_010 |
0.0921971318 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 3 |
Hb_002613_030 |
0.0957744236 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000668_190 |
0.1089967126 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 5 |
Hb_000164_030 |
0.11158559 |
- |
- |
PREDICTED: RING-H2 finger protein ATL14-like [Zea mays] |
| 6 |
Hb_006142_020 |
0.1116347865 |
- |
- |
JHL25P11.12 [Jatropha curcas] |
| 7 |
Hb_000739_250 |
0.1132966821 |
- |
- |
PREDICTED: uncharacterized protein LOC105120821 [Populus euphratica] |
| 8 |
Hb_000157_060 |
0.11341453 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004724_250 |
0.1153389706 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_048141_060 |
0.1270114414 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 11 |
Hb_144649_010 |
0.1312312699 |
- |
- |
60S ribosomal protein L34A [Hevea brasiliensis] |
| 12 |
Hb_002534_040 |
0.1345128701 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 13 |
Hb_000413_220 |
0.1384823869 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 14 |
Hb_108744_010 |
0.1425078106 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 15 |
Hb_028187_010 |
0.1440449023 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 16 |
Hb_169136_010 |
0.1481305729 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005649_070 |
0.1512921842 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004800_030 |
0.1516167188 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 19 |
Hb_074660_010 |
0.1519656211 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 20 |
Hb_000256_170 |
0.1521519048 |
- |
- |
PREDICTED: uncharacterized protein LOC105638320 [Jatropha curcas] |