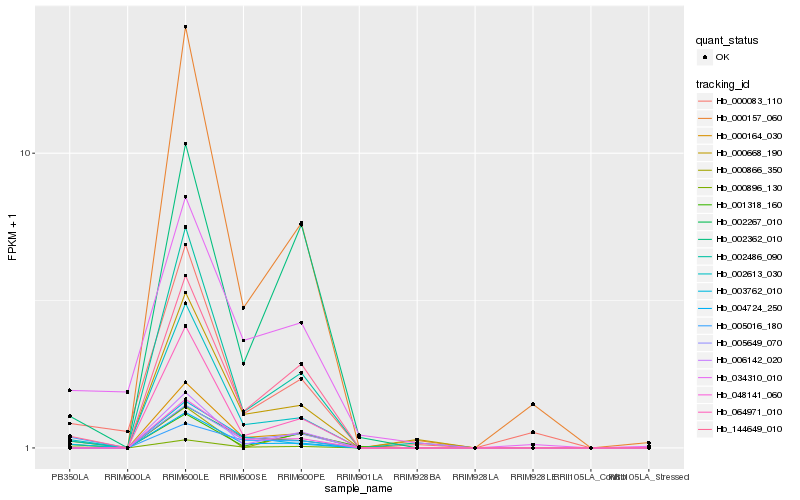
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000896_130 |
0.0 |
transcription factor |
TF Family: CPP |
cysteine-rich polycomb-like family protein [Populus trichocarpa] |
| 2 |
Hb_002613_030 |
0.1159662873 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_144649_010 |
0.1466158237 |
- |
- |
60S ribosomal protein L34A [Hevea brasiliensis] |
| 4 |
Hb_003762_010 |
0.1635123947 |
- |
- |
- |
| 5 |
Hb_002486_090 |
0.1683817 |
- |
- |
hypothetical protein POPTR_0005s06910g [Populus trichocarpa] |
| 6 |
Hb_000083_110 |
0.1814271436 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 7 |
Hb_001318_160 |
0.1925057299 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 8 |
Hb_000164_030 |
0.2055563648 |
- |
- |
PREDICTED: RING-H2 finger protein ATL14-like [Zea mays] |
| 9 |
Hb_064971_010 |
0.208622727 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 10 |
Hb_048141_060 |
0.2107249779 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 11 |
Hb_004724_250 |
0.2111120733 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 12 |
Hb_005016_180 |
0.2130918603 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 13 |
Hb_000866_350 |
0.2143707421 |
- |
- |
- |
| 14 |
Hb_002362_010 |
0.2150699611 |
- |
- |
hypothetical protein JCGZ_12328 [Jatropha curcas] |
| 15 |
Hb_006142_020 |
0.2197451148 |
- |
- |
JHL25P11.12 [Jatropha curcas] |
| 16 |
Hb_034310_010 |
0.2209541681 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_000157_060 |
0.2220869185 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005649_070 |
0.2233235748 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000668_190 |
0.2295746524 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 20 |
Hb_002267_010 |
0.2308870741 |
- |
- |
hypothetical protein SORBIDRAFT_01g040370 [Sorghum bicolor] |