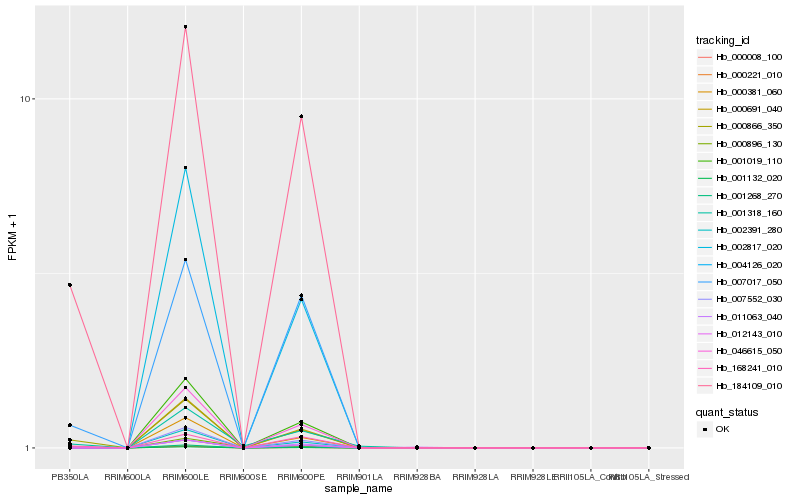
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000866_350 |
0.0 |
- |
- |
- |
| 2 |
Hb_184109_010 |
0.088152433 |
- |
- |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 3 |
Hb_168241_010 |
0.1726484566 |
- |
- |
hypothetical protein POPTR_0013s14620g, partial [Populus trichocarpa] |
| 4 |
Hb_007017_050 |
0.1759105423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001318_160 |
0.1762244988 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 6 |
Hb_011063_040 |
0.1829204336 |
- |
- |
PREDICTED: uncharacterized protein LOC100853799 [Vitis vinifera] |
| 7 |
Hb_000896_130 |
0.2143707421 |
transcription factor |
TF Family: CPP |
cysteine-rich polycomb-like family protein [Populus trichocarpa] |
| 8 |
Hb_012143_010 |
0.2245949646 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Vitis vinifera] |
| 9 |
Hb_001132_020 |
0.2246113091 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 10 |
Hb_046615_050 |
0.2246205355 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 11 |
Hb_000691_040 |
0.2246364392 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 12 |
Hb_001268_270 |
0.2246453125 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_000381_060 |
0.2246614579 |
- |
- |
- |
| 14 |
Hb_001019_110 |
0.2246721204 |
- |
- |
PREDICTED: cytochrome P450 734A1 [Jatropha curcas] |
| 15 |
Hb_004126_020 |
0.2247464261 |
- |
- |
PREDICTED: uncharacterized protein LOC105649549 [Jatropha curcas] |
| 16 |
Hb_000221_010 |
0.2247659542 |
- |
- |
polyprotein [Oryza australiensis] |
| 17 |
Hb_002817_020 |
0.2249629206 |
- |
- |
- |
| 18 |
Hb_007552_030 |
0.2249674648 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21064 [Jatropha curcas] |
| 19 |
Hb_002391_280 |
0.2249837916 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 20 |
Hb_000008_100 |
0.2250482022 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |