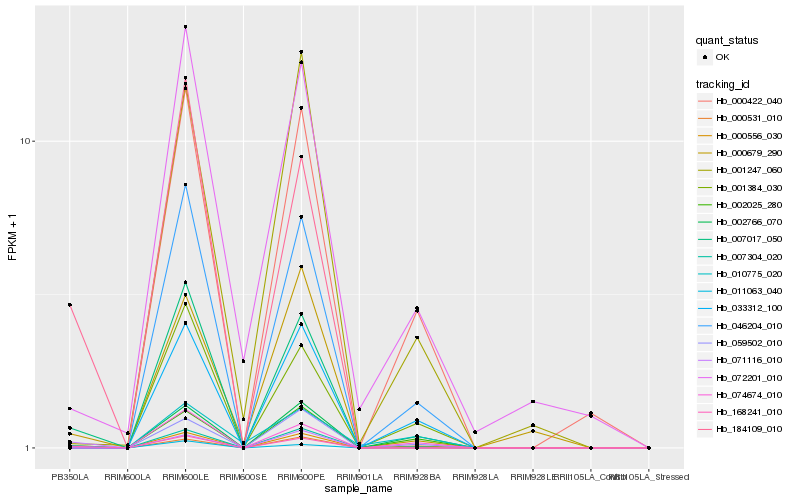
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000556_030 |
0.0 |
- |
- |
PREDICTED: probable carboxylesterase 9 [Jatropha curcas] |
| 2 |
Hb_011063_040 |
0.1268556452 |
- |
- |
PREDICTED: uncharacterized protein LOC100853799 [Vitis vinifera] |
| 3 |
Hb_168241_010 |
0.167593394 |
- |
- |
hypothetical protein POPTR_0013s14620g, partial [Populus trichocarpa] |
| 4 |
Hb_007017_050 |
0.1865223029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_033312_100 |
0.1899696521 |
- |
- |
- |
| 6 |
Hb_046204_010 |
0.1980977812 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 7 |
Hb_000531_010 |
0.1984270164 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0018s14830g [Populus trichocarpa] |
| 8 |
Hb_184109_010 |
0.2002245689 |
- |
- |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 9 |
Hb_002025_280 |
0.2007743016 |
- |
- |
PREDICTED: uncharacterized protein LOC105634201 [Jatropha curcas] |
| 10 |
Hb_000422_040 |
0.2027679062 |
- |
- |
hypothetical protein CICLE_v10013147mg [Citrus clementina] |
| 11 |
Hb_007304_020 |
0.2036088834 |
- |
- |
PREDICTED: uncharacterized protein LOC100256879 [Vitis vinifera] |
| 12 |
Hb_002766_070 |
0.2037359042 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |
| 13 |
Hb_059502_010 |
0.2054512607 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 14 |
Hb_001384_030 |
0.2085787339 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 15 |
Hb_000679_290 |
0.208633089 |
transcription factor |
TF Family: AUX/IAA |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_074674_010 |
0.208740616 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 17 |
Hb_001247_060 |
0.2115499061 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_071116_010 |
0.212547969 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 19 |
Hb_010775_020 |
0.2154438897 |
transcription factor |
TF Family: ERF |
Floral homeotic protein APETALA2, putative [Ricinus communis] |
| 20 |
Hb_072201_010 |
0.2176479252 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |