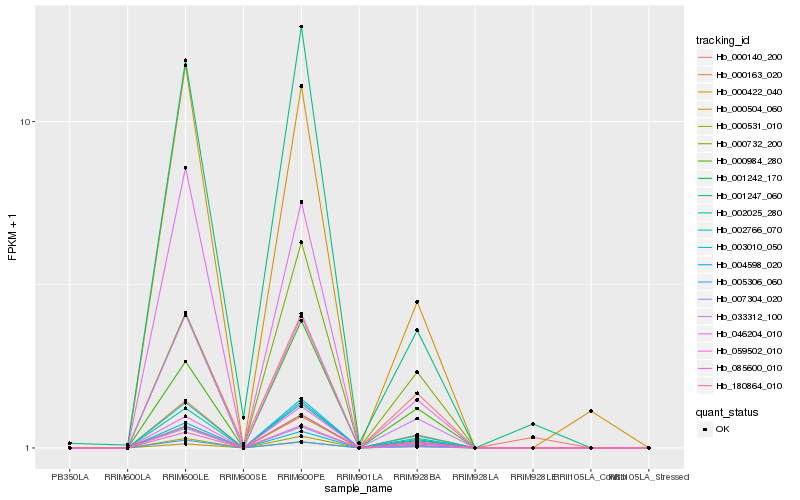
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002025_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634201 [Jatropha curcas] |
| 2 |
Hb_002766_070 |
0.0152566666 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |
| 3 |
Hb_033312_100 |
0.0465593594 |
- |
- |
- |
| 4 |
Hb_000531_010 |
0.0471725683 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0018s14830g [Populus trichocarpa] |
| 5 |
Hb_085600_010 |
0.05863216 |
- |
- |
PREDICTED: peroxidase 24-like [Jatropha curcas] |
| 6 |
Hb_001242_170 |
0.0709938063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_180864_010 |
0.0710088136 |
- |
- |
hypothetical protein B456_010G097000 [Gossypium raimondii] |
| 8 |
Hb_059502_010 |
0.0894978128 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 9 |
Hb_000984_280 |
0.0927495333 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 [Jatropha curcas] |
| 10 |
Hb_000504_060 |
0.1013274655 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 11 |
Hb_000422_040 |
0.1028230658 |
- |
- |
hypothetical protein CICLE_v10013147mg [Citrus clementina] |
| 12 |
Hb_005306_060 |
0.1102199368 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 13 |
Hb_004598_020 |
0.1238108208 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 14 |
Hb_000732_200 |
0.1246728465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000163_020 |
0.1273878763 |
- |
- |
- |
| 16 |
Hb_003010_050 |
0.1282259808 |
- |
- |
- |
| 17 |
Hb_007304_020 |
0.1286357718 |
- |
- |
PREDICTED: uncharacterized protein LOC100256879 [Vitis vinifera] |
| 18 |
Hb_001247_060 |
0.1313786651 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_000140_200 |
0.1313790689 |
- |
- |
PREDICTED: uncharacterized protein LOC105642928 [Jatropha curcas] |
| 20 |
Hb_046204_010 |
0.1313893823 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |