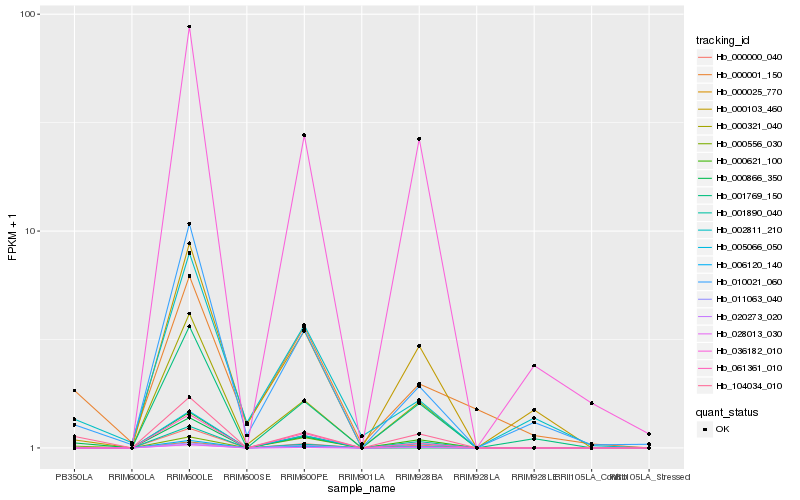
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_104034_010 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Pyrus x bretschneideri] |
| 2 |
Hb_011063_040 |
0.1737321941 |
- |
- |
PREDICTED: uncharacterized protein LOC100853799 [Vitis vinifera] |
| 3 |
Hb_010021_060 |
0.2384717201 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 4 |
Hb_006120_140 |
0.2404522951 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 5 |
Hb_000621_100 |
0.2416472259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000321_040 |
0.2481019746 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 7 |
Hb_000000_040 |
0.2508603019 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like [Jatropha curcas] |
| 8 |
Hb_001890_040 |
0.2511800171 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 9 |
Hb_020273_020 |
0.2524928887 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 10 |
Hb_028013_030 |
0.2564236225 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_036182_010 |
0.2575120481 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |
| 12 |
Hb_000001_150 |
0.2597249222 |
- |
- |
PREDICTED: uncharacterized protein LOC105138186 isoform X1 [Populus euphratica] |
| 13 |
Hb_001769_150 |
0.2629192848 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 14 |
Hb_005066_050 |
0.2643633033 |
- |
- |
- |
| 15 |
Hb_000025_770 |
0.2655408239 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 16 |
Hb_061361_010 |
0.2668238146 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_000103_460 |
0.2669566807 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 18 |
Hb_000556_030 |
0.2687443179 |
- |
- |
PREDICTED: probable carboxylesterase 9 [Jatropha curcas] |
| 19 |
Hb_002811_210 |
0.2716459929 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000866_350 |
0.2738781396 |
- |
- |
- |