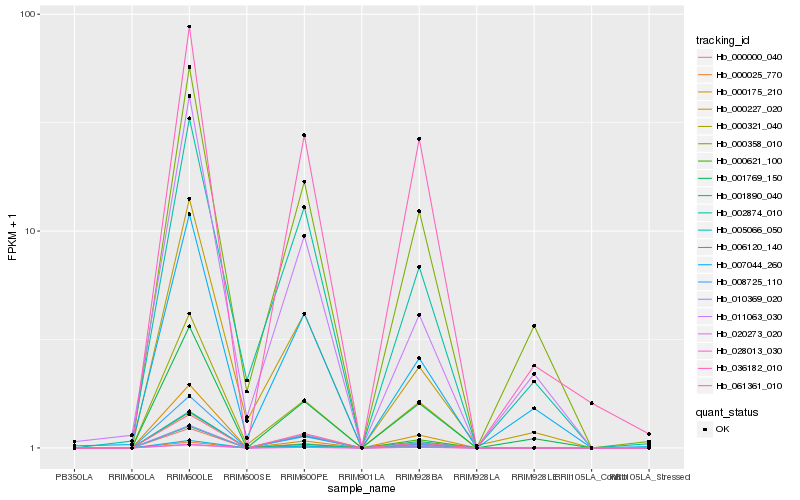
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006120_140 |
0.0 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 2 |
Hb_000621_100 |
0.0465388695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000000_040 |
0.0510536483 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like [Jatropha curcas] |
| 4 |
Hb_020273_020 |
0.0587966205 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_000321_040 |
0.0638645029 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 6 |
Hb_001890_040 |
0.0820531009 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 7 |
Hb_005066_050 |
0.1014078054 |
- |
- |
- |
| 8 |
Hb_028013_030 |
0.1031907336 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_000025_770 |
0.1148540603 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 10 |
Hb_036182_010 |
0.116657986 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |
| 11 |
Hb_001769_150 |
0.1180377578 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 12 |
Hb_061361_010 |
0.1189473802 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_000175_210 |
0.1314447028 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 14 |
Hb_008725_110 |
0.1386676642 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000227_020 |
0.1452732503 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 16 |
Hb_000358_010 |
0.1486539767 |
- |
- |
PREDICTED: basic blue protein [Populus euphratica] |
| 17 |
Hb_010369_020 |
0.158169654 |
- |
- |
hypothetical protein JCGZ_21088 [Jatropha curcas] |
| 18 |
Hb_007044_260 |
0.1588846803 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 19 |
Hb_011063_030 |
0.1684192173 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_002874_010 |
0.1690899574 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit A isoform X1 [Jatropha curcas] |