| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
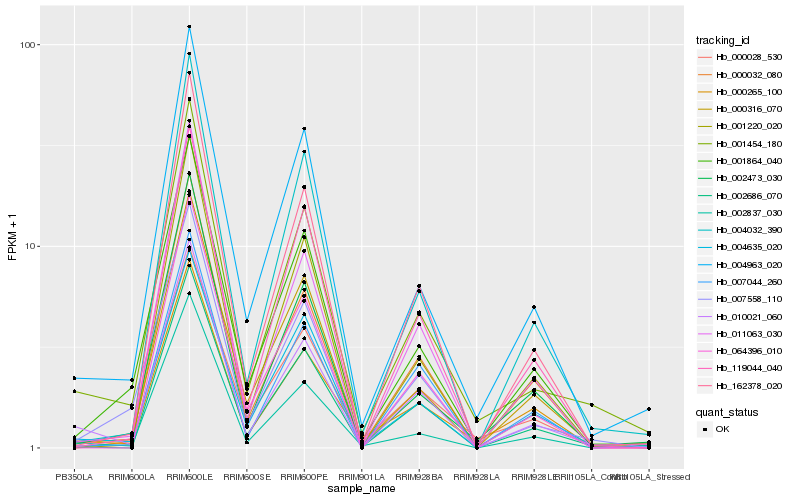
Hb_010021_060 |
0.0 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001454_180 |
0.0941481857 |
- |
- |
PREDICTED: uncharacterized protein LOC105643432 [Jatropha curcas] |
| 3 |
Hb_002837_030 |
0.1054961707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_162378_020 |
0.1065494638 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 13-like [Populus euphratica] |
| 5 |
Hb_011063_030 |
0.1065893454 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_002686_070 |
0.1088892612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007044_260 |
0.1092857467 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004963_020 |
0.1098532863 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 9 |
Hb_004032_390 |
0.1173810639 |
- |
- |
CDK, putative [Ricinus communis] |
| 10 |
Hb_000032_080 |
0.121805828 |
transcription factor |
TF Family: HMG |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_004635_020 |
0.122612103 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 12 |
Hb_000028_530 |
0.1229996088 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 13 |
Hb_007558_110 |
0.1250823255 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 14 |
Hb_000316_070 |
0.1253808059 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 15 |
Hb_002473_030 |
0.1257323305 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 16 |
Hb_064396_010 |
0.1259041941 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000265_100 |
0.1269578778 |
- |
- |
PREDICTED: uncharacterized protein LOC105643473 [Jatropha curcas] |
| 18 |
Hb_001220_020 |
0.1288408342 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Jatropha curcas] |
| 19 |
Hb_001864_040 |
0.1315087546 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 20 |
Hb_119044_040 |
0.1317457759 |
- |
- |
cyclin B, putative [Ricinus communis] |