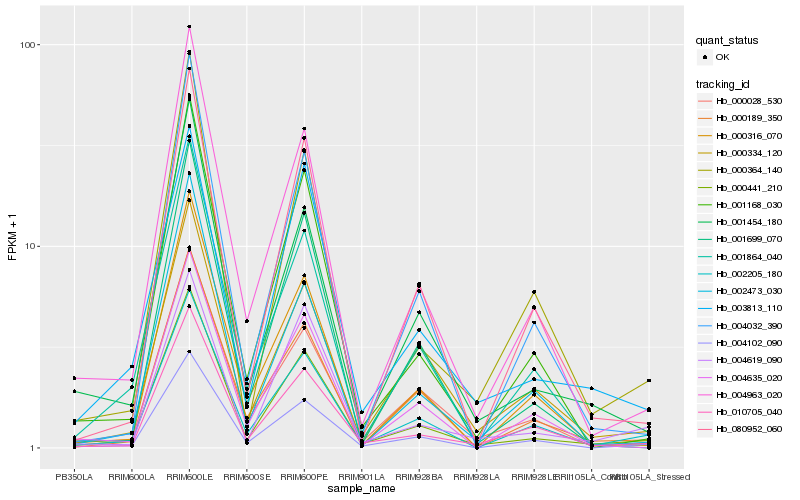
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000441_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 2 |
Hb_004963_020 |
0.0907352782 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 3 |
Hb_004102_090 |
0.0914426682 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 4 |
Hb_010705_040 |
0.0944034118 |
- |
- |
PREDICTED: kinesin-4 [Jatropha curcas] |
| 5 |
Hb_001454_180 |
0.094639493 |
- |
- |
PREDICTED: uncharacterized protein LOC105643432 [Jatropha curcas] |
| 6 |
Hb_002205_180 |
0.0947488987 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 7 |
Hb_001168_030 |
0.0990493642 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 8 |
Hb_001864_040 |
0.1080350255 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 9 |
Hb_000028_530 |
0.109241711 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 10 |
Hb_080952_060 |
0.1096817852 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 11 |
Hb_003813_110 |
0.1117734308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001699_070 |
0.114643413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000189_350 |
0.1148320545 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 3 [Jatropha curcas] |
| 14 |
Hb_004619_090 |
0.1158389116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004032_390 |
0.1171900731 |
- |
- |
CDK, putative [Ricinus communis] |
| 16 |
Hb_000316_070 |
0.1174738846 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 17 |
Hb_000364_140 |
0.1178637391 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 18 |
Hb_002473_030 |
0.1193226906 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 19 |
Hb_000334_120 |
0.1215765861 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 20 |
Hb_004635_020 |
0.12263099 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |