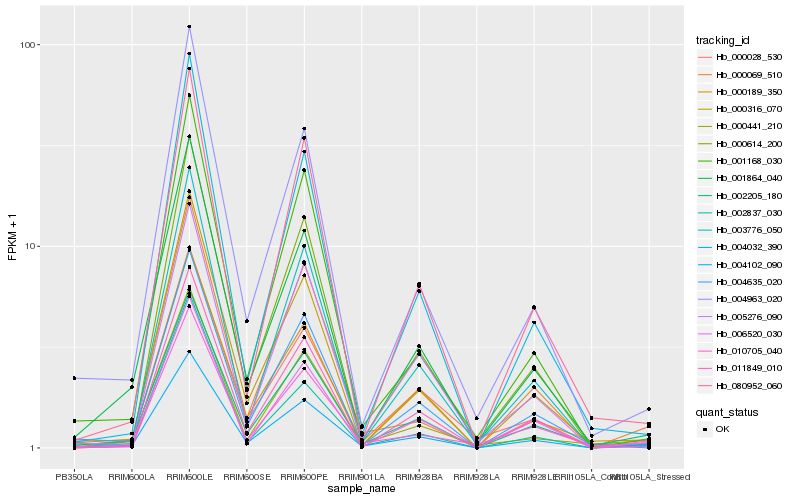
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004102_090 |
0.0 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 2 |
Hb_002205_180 |
0.0519986321 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 3 |
Hb_004963_020 |
0.0884670109 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 4 |
Hb_000441_210 |
0.0914426682 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 5 |
Hb_000316_070 |
0.0928945279 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 6 |
Hb_006520_030 |
0.0932004805 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 7 |
Hb_000189_350 |
0.0960223748 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 3 [Jatropha curcas] |
| 8 |
Hb_001864_040 |
0.0972010024 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 9 |
Hb_010705_040 |
0.0985251871 |
- |
- |
PREDICTED: kinesin-4 [Jatropha curcas] |
| 10 |
Hb_000028_530 |
0.0986703527 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 11 |
Hb_001168_030 |
0.0990539346 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 12 |
Hb_004635_020 |
0.1022185877 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 13 |
Hb_011849_010 |
0.1029668946 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 14 |
Hb_000069_510 |
0.1047052865 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 15 |
Hb_002837_030 |
0.111023628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_080952_060 |
0.112866835 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 17 |
Hb_003776_050 |
0.1130541707 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 18 |
Hb_004032_390 |
0.1131438832 |
- |
- |
CDK, putative [Ricinus communis] |
| 19 |
Hb_005276_090 |
0.1131562092 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000614_200 |
0.1140583567 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |