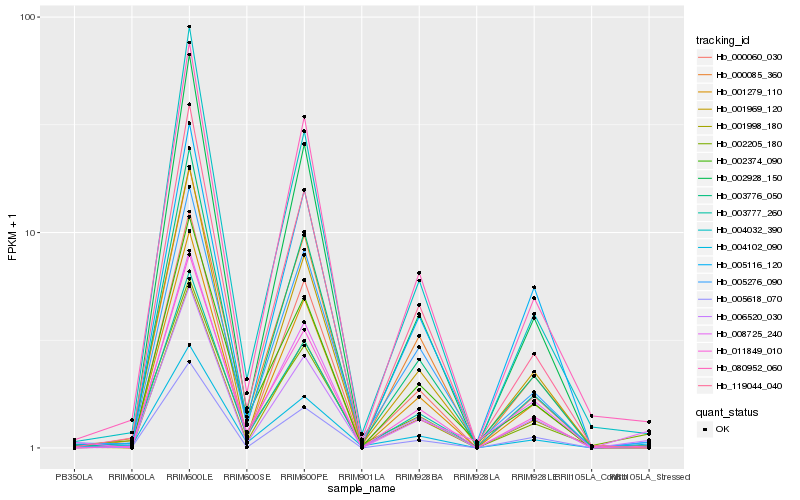
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006520_030 |
0.0 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 2 |
Hb_011849_010 |
0.0776794381 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 3 |
Hb_001998_180 |
0.0829428452 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 4 |
Hb_003776_050 |
0.0849249252 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 5 |
Hb_080952_060 |
0.0861162956 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 6 |
Hb_001969_120 |
0.0876863145 |
- |
- |
PREDICTED: plectin [Jatropha curcas] |
| 7 |
Hb_003777_260 |
0.091829505 |
- |
- |
PREDICTED: uncharacterized protein LOC105640927 isoform X4 [Jatropha curcas] |
| 8 |
Hb_001279_110 |
0.09212668 |
- |
- |
PREDICTED: uncharacterized protein LOC105633338 isoform X4 [Jatropha curcas] |
| 9 |
Hb_000060_030 |
0.0928257962 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like [Jatropha curcas] |
| 10 |
Hb_004102_090 |
0.0932004805 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 11 |
Hb_000085_360 |
0.094690737 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 12 |
Hb_005618_070 |
0.0956869701 |
- |
- |
PREDICTED: uncharacterized protein LOC105643318 [Jatropha curcas] |
| 13 |
Hb_119044_040 |
0.0957182735 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 14 |
Hb_004032_390 |
0.0972084638 |
- |
- |
CDK, putative [Ricinus communis] |
| 15 |
Hb_008725_240 |
0.0988066744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005276_090 |
0.0988288159 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002205_180 |
0.0994596793 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 18 |
Hb_005116_120 |
0.0999593823 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002374_090 |
0.1010861982 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 20 |
Hb_002928_150 |
0.1011126364 |
- |
- |
PREDICTED: cyclin-B2-4-like [Jatropha curcas] |