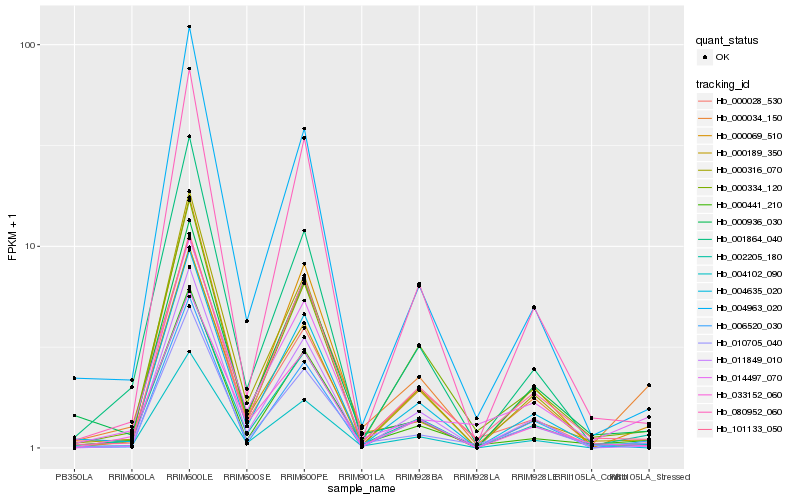
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002205_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 2 |
Hb_004102_090 |
0.0519986321 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 3 |
Hb_000189_350 |
0.0828764898 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 3 [Jatropha curcas] |
| 4 |
Hb_000441_210 |
0.0947488987 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 5 |
Hb_010705_040 |
0.0993492179 |
- |
- |
PREDICTED: kinesin-4 [Jatropha curcas] |
| 6 |
Hb_006520_030 |
0.0994596793 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 7 |
Hb_033152_060 |
0.102123978 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 [Jatropha curcas] |
| 8 |
Hb_000028_530 |
0.105236554 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 9 |
Hb_000316_070 |
0.1055388596 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 10 |
Hb_000334_120 |
0.1070007071 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 11 |
Hb_004635_020 |
0.1074483062 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 12 |
Hb_001864_040 |
0.1098495099 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 13 |
Hb_004963_020 |
0.1114237426 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 14 |
Hb_080952_060 |
0.1133877551 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 15 |
Hb_101133_050 |
0.1134035811 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 16 |
Hb_011849_010 |
0.1173731345 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 17 |
Hb_000034_150 |
0.1177085993 |
- |
- |
PREDICTED: kinesin-like protein BC2 [Jatropha curcas] |
| 18 |
Hb_000936_030 |
0.1185321185 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 19 |
Hb_000069_510 |
0.1199001814 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 20 |
Hb_014497_070 |
0.1200300324 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |