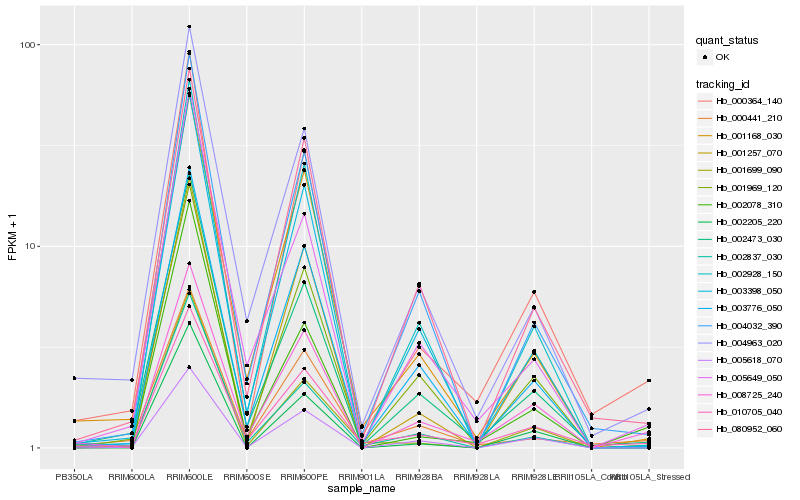
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000364_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 2 |
Hb_002473_030 |
0.0859956169 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 3 |
Hb_010705_040 |
0.0941907844 |
- |
- |
PREDICTED: kinesin-4 [Jatropha curcas] |
| 4 |
Hb_004963_020 |
0.1087472355 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 5 |
Hb_004032_390 |
0.1108360521 |
- |
- |
CDK, putative [Ricinus communis] |
| 6 |
Hb_001699_090 |
0.1146780254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001168_030 |
0.1153775088 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 8 |
Hb_002078_310 |
0.1165867742 |
- |
- |
PREDICTED: uncharacterized protein LOC105645005 [Jatropha curcas] |
| 9 |
Hb_003398_050 |
0.1170671695 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |
| 10 |
Hb_008725_240 |
0.117139157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000441_210 |
0.1178637391 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 12 |
Hb_080952_060 |
0.1181310435 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 13 |
Hb_002928_150 |
0.1182993092 |
- |
- |
PREDICTED: cyclin-B2-4-like [Jatropha curcas] |
| 14 |
Hb_001969_120 |
0.1185821646 |
- |
- |
PREDICTED: plectin [Jatropha curcas] |
| 15 |
Hb_002837_030 |
0.120145091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003776_050 |
0.1210273098 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 17 |
Hb_001257_070 |
0.1225872918 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1-like [Jatropha curcas] |
| 18 |
Hb_005649_050 |
0.1236139658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002205_220 |
0.1243104042 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 20 |
Hb_005618_070 |
0.1247571129 |
- |
- |
PREDICTED: uncharacterized protein LOC105643318 [Jatropha curcas] |