| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
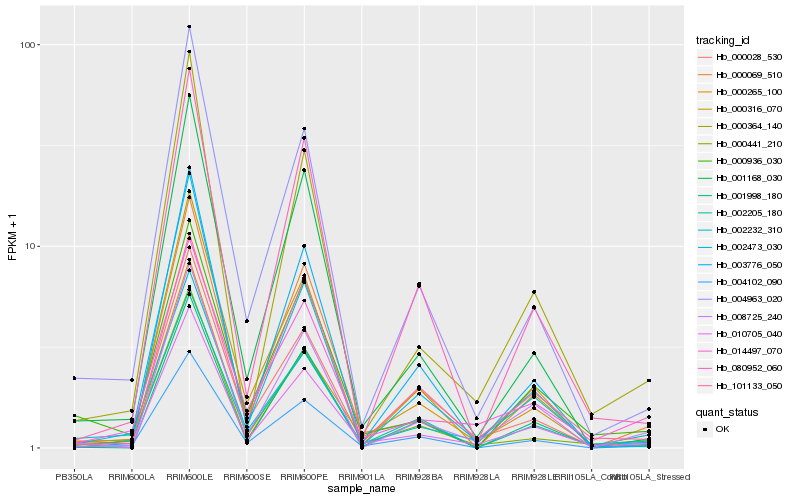
Hb_010705_040 |
0.0 |
- |
- |
PREDICTED: kinesin-4 [Jatropha curcas] |
| 2 |
Hb_000364_140 |
0.0941907844 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 3 |
Hb_000441_210 |
0.0944034118 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 4 |
Hb_004963_020 |
0.0977606395 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 5 |
Hb_004102_090 |
0.0985251871 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 6 |
Hb_002205_180 |
0.0993492179 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 7 |
Hb_000265_100 |
0.0994270303 |
- |
- |
PREDICTED: uncharacterized protein LOC105643473 [Jatropha curcas] |
| 8 |
Hb_000069_510 |
0.0994665605 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 9 |
Hb_002232_310 |
0.1007276654 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 10 |
Hb_001168_030 |
0.1011398666 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 11 |
Hb_014497_070 |
0.1034584561 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |
| 12 |
Hb_008725_240 |
0.1053371365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000316_070 |
0.106820374 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 14 |
Hb_101133_050 |
0.1075905823 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 15 |
Hb_002473_030 |
0.1080933187 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 16 |
Hb_000028_530 |
0.11119865 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 17 |
Hb_080952_060 |
0.1121599202 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 18 |
Hb_000936_030 |
0.113479546 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 19 |
Hb_001998_180 |
0.1158750784 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 20 |
Hb_003776_050 |
0.1160122551 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |