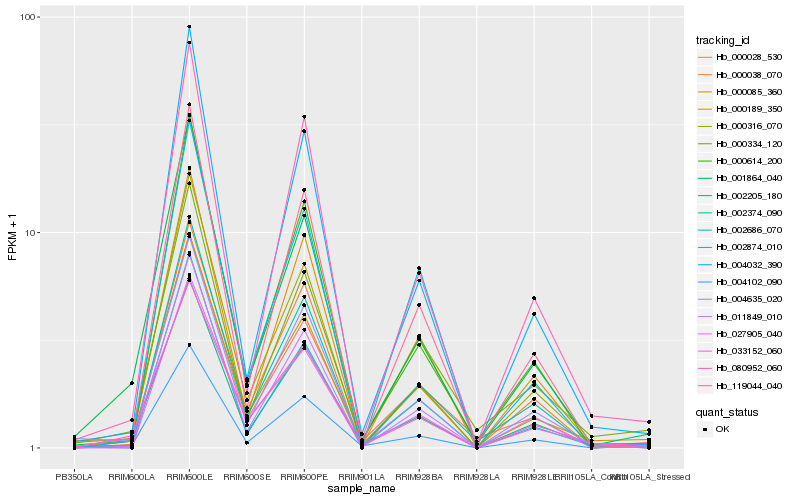
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_350 |
0.0 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 3 [Jatropha curcas] |
| 2 |
Hb_033152_060 |
0.0771216013 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 [Jatropha curcas] |
| 3 |
Hb_002205_180 |
0.0828764898 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 4 |
Hb_000316_070 |
0.0844259057 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 5 |
Hb_027905_040 |
0.0889854168 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: E2F transcription factor-like E2FF [Jatropha curcas] |
| 6 |
Hb_011849_010 |
0.0901068574 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 7 |
Hb_000028_530 |
0.0903729919 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 8 |
Hb_000334_120 |
0.0910269149 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 9 |
Hb_002374_090 |
0.0950993986 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 10 |
Hb_004102_090 |
0.0960223748 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 11 |
Hb_004635_020 |
0.0967518389 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 12 |
Hb_002686_070 |
0.097581306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000085_360 |
0.0977251062 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 14 |
Hb_000038_070 |
0.0997336074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001864_040 |
0.1020915289 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 16 |
Hb_080952_060 |
0.1034349763 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 17 |
Hb_119044_040 |
0.1067201247 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 18 |
Hb_004032_390 |
0.1067584064 |
- |
- |
CDK, putative [Ricinus communis] |
| 19 |
Hb_002874_010 |
0.1096092742 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit A isoform X1 [Jatropha curcas] |
| 20 |
Hb_000614_200 |
0.1097695538 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |