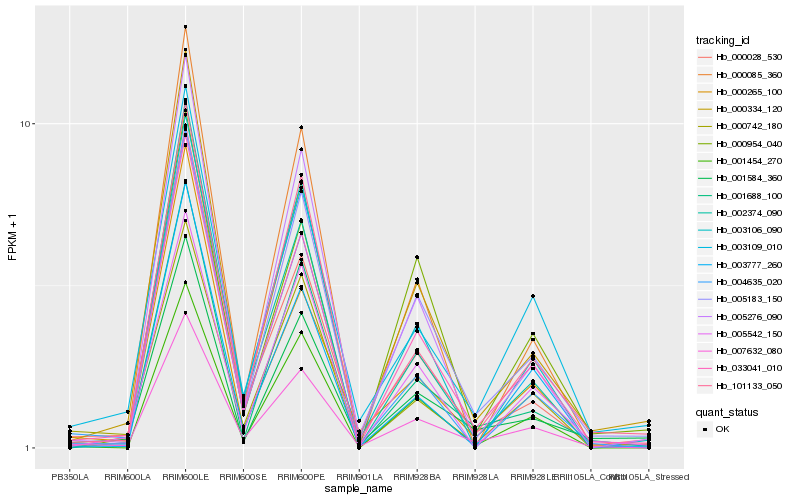
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033041_010 |
0.0 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2-like [Populus euphratica] |
| 2 |
Hb_007632_080 |
0.0558020433 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 3 |
Hb_000334_120 |
0.0718949806 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 4 |
Hb_005183_150 |
0.0822871773 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 5 |
Hb_101133_050 |
0.0931608734 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 6 |
Hb_000028_530 |
0.0934587421 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 7 |
Hb_005276_090 |
0.0990403343 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003109_010 |
0.0993120172 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 9 |
Hb_002374_090 |
0.1012347474 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 10 |
Hb_005542_150 |
0.1016329639 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 11 |
Hb_001454_270 |
0.1040477573 |
- |
- |
PREDICTED: condensin-2 complex subunit D3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001688_100 |
0.1047742779 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 13 |
Hb_001584_360 |
0.1051374043 |
- |
- |
PREDICTED: serine/threonine-protein kinase haspin homolog hrk1 [Jatropha curcas] |
| 14 |
Hb_000085_360 |
0.1060654073 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 15 |
Hb_003106_090 |
0.1074310551 |
transcription factor |
TF Family: E2F-DP |
E2F, putative [Ricinus communis] |
| 16 |
Hb_000954_040 |
0.1128668981 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 17 |
Hb_003777_260 |
0.1156512157 |
- |
- |
PREDICTED: uncharacterized protein LOC105640927 isoform X4 [Jatropha curcas] |
| 18 |
Hb_000265_100 |
0.1166251689 |
- |
- |
PREDICTED: uncharacterized protein LOC105643473 [Jatropha curcas] |
| 19 |
Hb_000742_180 |
0.117570539 |
- |
- |
hypothetical protein POPTR_0011s16640g [Populus trichocarpa] |
| 20 |
Hb_004635_020 |
0.1177420385 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |