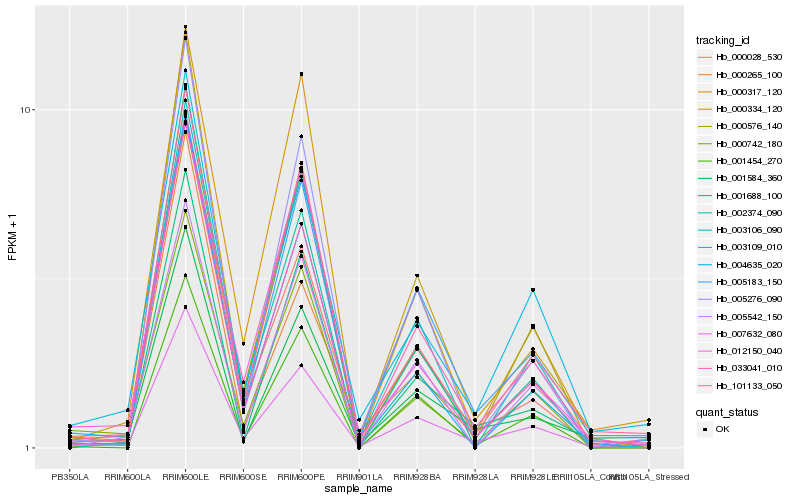
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007632_080 |
0.0 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 2 |
Hb_033041_010 |
0.0558020433 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2-like [Populus euphratica] |
| 3 |
Hb_005183_150 |
0.0984727798 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 4 |
Hb_003109_010 |
0.1008449388 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 5 |
Hb_000028_530 |
0.1028214465 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 6 |
Hb_101133_050 |
0.1031802316 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 7 |
Hb_000334_120 |
0.1064565073 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 8 |
Hb_000576_140 |
0.113688465 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 9 |
Hb_001584_360 |
0.1142042125 |
- |
- |
PREDICTED: serine/threonine-protein kinase haspin homolog hrk1 [Jatropha curcas] |
| 10 |
Hb_000742_180 |
0.1144114958 |
- |
- |
hypothetical protein POPTR_0011s16640g [Populus trichocarpa] |
| 11 |
Hb_004635_020 |
0.1147396792 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 12 |
Hb_005276_090 |
0.1151549959 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002374_090 |
0.1171645345 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 14 |
Hb_001454_270 |
0.1175164596 |
- |
- |
PREDICTED: condensin-2 complex subunit D3 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001688_100 |
0.1175410884 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 16 |
Hb_012150_040 |
0.1196636526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000265_100 |
0.1215382431 |
- |
- |
PREDICTED: uncharacterized protein LOC105643473 [Jatropha curcas] |
| 18 |
Hb_003106_090 |
0.122767781 |
transcription factor |
TF Family: E2F-DP |
E2F, putative [Ricinus communis] |
| 19 |
Hb_000317_120 |
0.1231165455 |
- |
- |
PREDICTED: uncharacterized protein LOC105647000 [Jatropha curcas] |
| 20 |
Hb_005542_150 |
0.125572546 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |