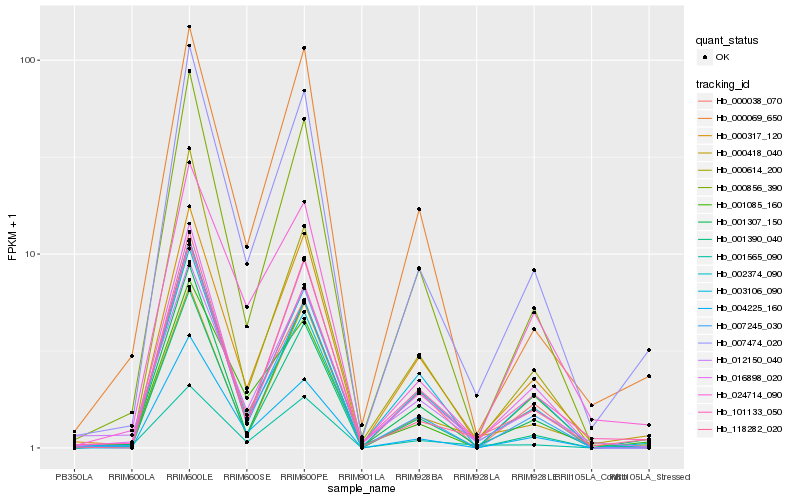
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007474_020 |
0.0 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 2 |
Hb_101133_050 |
0.085758225 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 3 |
Hb_001390_040 |
0.0950398786 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 4 |
Hb_000856_390 |
0.0973043911 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001307_150 |
0.0974913163 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000317_120 |
0.0980233836 |
- |
- |
PREDICTED: uncharacterized protein LOC105647000 [Jatropha curcas] |
| 7 |
Hb_118282_020 |
0.098241509 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X3 [Jatropha curcas] |
| 8 |
Hb_000038_070 |
0.1002123407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_016898_020 |
0.1041285569 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105120899 [Populus euphratica] |
| 10 |
Hb_000418_040 |
0.1075028116 |
- |
- |
hypothetical protein RCOM_0377590 [Ricinus communis] |
| 11 |
Hb_004225_160 |
0.1118533025 |
- |
- |
PREDICTED: kiwellin-like [Jatropha curcas] |
| 12 |
Hb_000069_650 |
0.1131402162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001565_090 |
0.1141410607 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003106_090 |
0.1148003357 |
transcription factor |
TF Family: E2F-DP |
E2F, putative [Ricinus communis] |
| 15 |
Hb_001085_160 |
0.1160946689 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 16 |
Hb_007245_030 |
0.1163959131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_012150_040 |
0.1165016355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002374_090 |
0.1169800872 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 19 |
Hb_024714_090 |
0.1212521005 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 20 |
Hb_000614_200 |
0.1212581961 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |