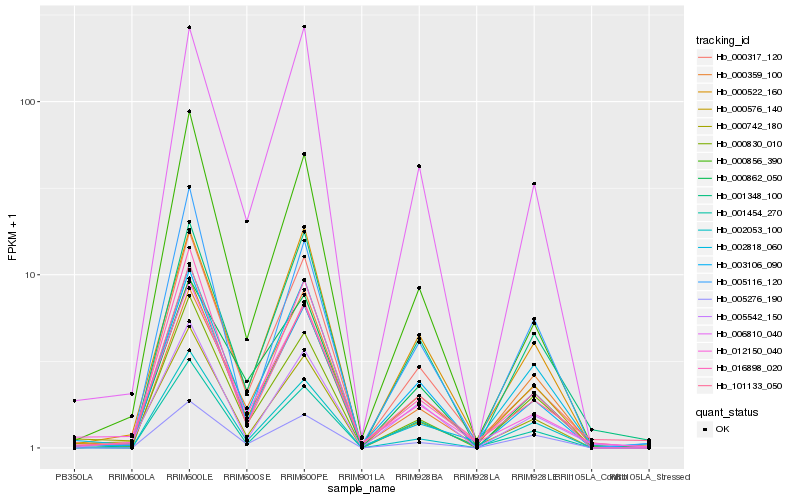
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000576_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 2 |
Hb_000317_120 |
0.0837545433 |
- |
- |
PREDICTED: uncharacterized protein LOC105647000 [Jatropha curcas] |
| 3 |
Hb_000742_180 |
0.0860750342 |
- |
- |
hypothetical protein POPTR_0011s16640g [Populus trichocarpa] |
| 4 |
Hb_000830_010 |
0.087946659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003106_090 |
0.0904517893 |
transcription factor |
TF Family: E2F-DP |
E2F, putative [Ricinus communis] |
| 6 |
Hb_016898_020 |
0.0909517825 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105120899 [Populus euphratica] |
| 7 |
Hb_005276_190 |
0.0957389926 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000522_160 |
0.100019551 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 9 |
Hb_006810_040 |
0.1049551842 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 10 |
Hb_001454_270 |
0.1058589719 |
- |
- |
PREDICTED: condensin-2 complex subunit D3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_101133_050 |
0.1073417222 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 12 |
Hb_012150_040 |
0.1074087432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002818_060 |
0.1094993682 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_005542_150 |
0.1101576857 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 15 |
Hb_000359_100 |
0.1106944063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000856_390 |
0.1117358751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000862_050 |
0.1118753618 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 18 |
Hb_005116_120 |
0.1118993937 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002053_100 |
0.1122154253 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 20 |
Hb_001348_100 |
0.1129981494 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |