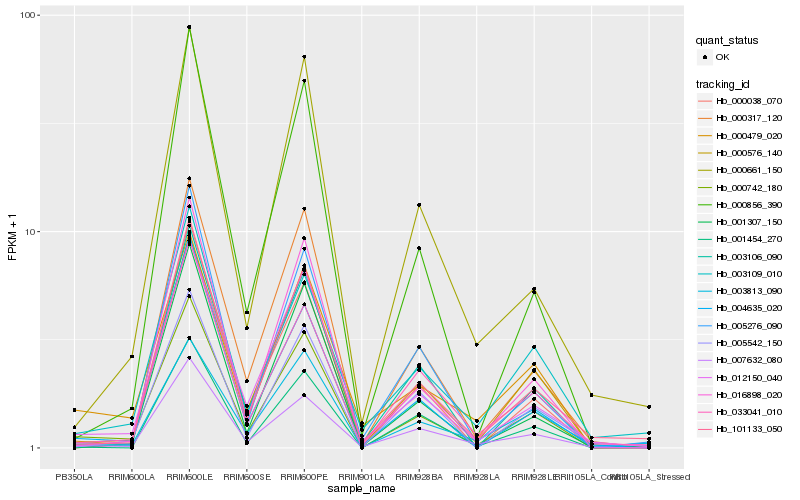
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000742_180 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s16640g [Populus trichocarpa] |
| 2 |
Hb_000576_140 |
0.0860750342 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 3 |
Hb_012150_040 |
0.1002093397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007632_080 |
0.1144114958 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 5 |
Hb_101133_050 |
0.1151857641 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 6 |
Hb_016898_020 |
0.1159967744 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105120899 [Populus euphratica] |
| 7 |
Hb_033041_010 |
0.117570539 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2-like [Populus euphratica] |
| 8 |
Hb_003106_090 |
0.1183292898 |
transcription factor |
TF Family: E2F-DP |
E2F, putative [Ricinus communis] |
| 9 |
Hb_000479_020 |
0.1194613385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000317_120 |
0.1217990282 |
- |
- |
PREDICTED: uncharacterized protein LOC105647000 [Jatropha curcas] |
| 11 |
Hb_004635_020 |
0.1229729176 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 12 |
Hb_005542_150 |
0.1273128303 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 13 |
Hb_000856_390 |
0.1286651996 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003109_010 |
0.1299528032 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 15 |
Hb_003813_090 |
0.1310710912 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 16 |
Hb_000038_070 |
0.1318163031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001307_150 |
0.1333581886 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_000661_150 |
0.1343483317 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 19 |
Hb_005276_090 |
0.1345509631 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001454_270 |
0.1360400717 |
- |
- |
PREDICTED: condensin-2 complex subunit D3 isoform X2 [Jatropha curcas] |