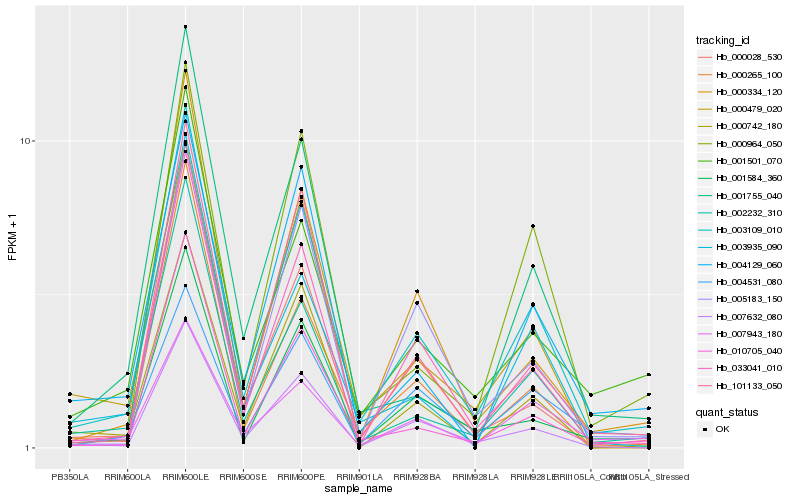
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003109_010 |
0.0 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 2 |
Hb_033041_010 |
0.0993120172 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2-like [Populus euphratica] |
| 3 |
Hb_007632_080 |
0.1008449388 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 4 |
Hb_002232_310 |
0.1046759151 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 5 |
Hb_001584_360 |
0.1179576525 |
- |
- |
PREDICTED: serine/threonine-protein kinase haspin homolog hrk1 [Jatropha curcas] |
| 6 |
Hb_101133_050 |
0.1191372354 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 7 |
Hb_007943_180 |
0.1204600642 |
- |
- |
PREDICTED: uncharacterized protein LOC105637734 [Jatropha curcas] |
| 8 |
Hb_000334_120 |
0.1224985479 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 9 |
Hb_000265_100 |
0.1251132595 |
- |
- |
PREDICTED: uncharacterized protein LOC105643473 [Jatropha curcas] |
| 10 |
Hb_001501_070 |
0.1268646808 |
- |
- |
PREDICTED: condensin complex subunit 2 [Jatropha curcas] |
| 11 |
Hb_004531_080 |
0.1277304681 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 12 |
Hb_000964_050 |
0.1293207987 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 13 |
Hb_000742_180 |
0.1299528032 |
- |
- |
hypothetical protein POPTR_0011s16640g [Populus trichocarpa] |
| 14 |
Hb_003935_090 |
0.130294582 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 15 |
Hb_001755_040 |
0.1308018214 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_010705_040 |
0.1308760379 |
- |
- |
PREDICTED: kinesin-4 [Jatropha curcas] |
| 17 |
Hb_000479_020 |
0.133838113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005183_150 |
0.1338832188 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 19 |
Hb_004129_060 |
0.1339153696 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 20 |
Hb_000028_530 |
0.1340132093 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |