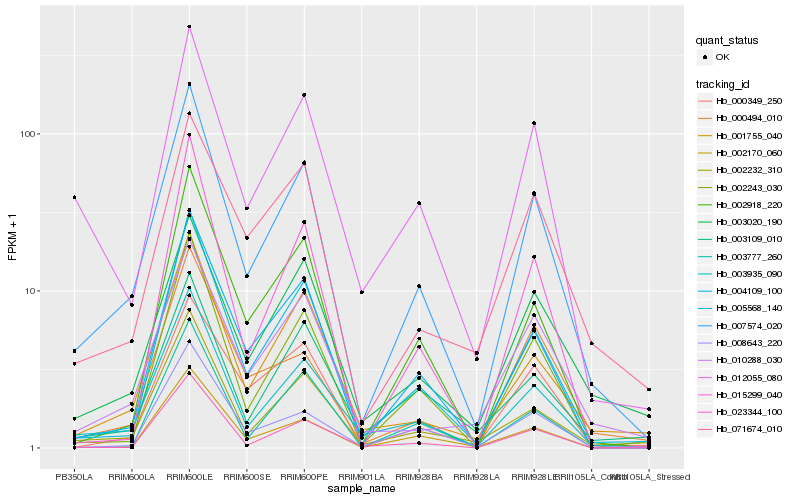
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007574_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 2 |
Hb_003935_090 |
0.081788438 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 3 |
Hb_004109_100 |
0.0989425183 |
- |
- |
Interactor of constitutive active rops 1 isoform 1 [Theobroma cacao] |
| 4 |
Hb_002232_310 |
0.1080943912 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 5 |
Hb_005568_140 |
0.1113557421 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_001755_040 |
0.1146175553 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002243_030 |
0.1190298383 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 3-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002918_220 |
0.1294550903 |
- |
- |
PREDICTED: uncharacterized protein LOC105138896 [Populus euphratica] |
| 9 |
Hb_002170_060 |
0.1316788276 |
- |
- |
PREDICTED: mitogen-activated protein kinase-binding protein 1 [Jatropha curcas] |
| 10 |
Hb_010288_030 |
0.1328138302 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 11 |
Hb_012055_080 |
0.1329431644 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 12 |
Hb_003020_190 |
0.138597796 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 13 |
Hb_008643_220 |
0.1387891079 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 14 |
Hb_023344_100 |
0.1396246802 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |
| 15 |
Hb_015299_040 |
0.1402722432 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 16 |
Hb_000349_250 |
0.1406551368 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 17 |
Hb_000494_010 |
0.1439988606 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0014s04460g [Populus trichocarpa] |
| 18 |
Hb_003109_010 |
0.1447201117 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 19 |
Hb_071674_010 |
0.1487439169 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 20 |
Hb_003777_260 |
0.1487685873 |
- |
- |
PREDICTED: uncharacterized protein LOC105640927 isoform X4 [Jatropha curcas] |