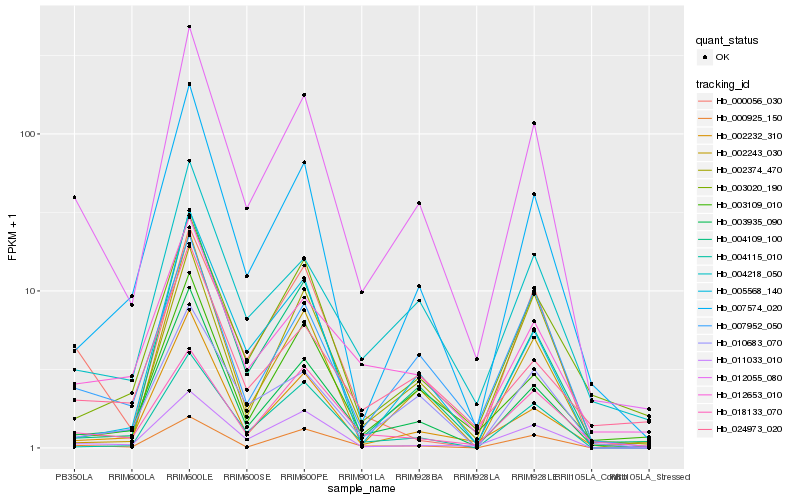
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012055_080 |
0.0 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 2 |
Hb_003935_090 |
0.1168895985 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 3 |
Hb_007952_050 |
0.1240334677 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein-like [Jatropha curcas] |
| 4 |
Hb_011033_010 |
0.1329316674 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 5 |
Hb_007574_020 |
0.1329431644 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 6 |
Hb_004218_050 |
0.1351370933 |
- |
- |
annexin [Manihot esculenta] |
| 7 |
Hb_000925_150 |
0.1391363866 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 8 |
Hb_012653_010 |
0.1481631114 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 9 |
Hb_004109_100 |
0.149563484 |
- |
- |
Interactor of constitutive active rops 1 isoform 1 [Theobroma cacao] |
| 10 |
Hb_002232_310 |
0.1525098242 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 11 |
Hb_003109_010 |
0.1537347278 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 12 |
Hb_010683_070 |
0.15381399 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004115_010 |
0.1561605774 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 14 |
Hb_024973_020 |
0.1578351349 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 15 |
Hb_003020_190 |
0.158551043 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 16 |
Hb_005568_140 |
0.1598474151 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_000056_030 |
0.1622838628 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g14390 isoform X1 [Jatropha curcas] |
| 18 |
Hb_018133_070 |
0.1650476314 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 19 |
Hb_002374_470 |
0.1651846099 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 20 |
Hb_002243_030 |
0.1655792569 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 3-like isoform X1 [Jatropha curcas] |