| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
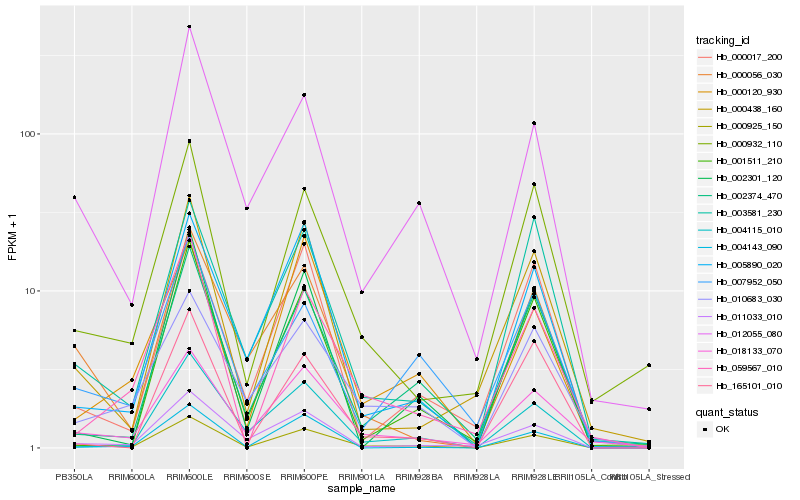
Hb_000925_150 |
0.0 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 2 |
Hb_018133_070 |
0.1287895235 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 3 |
Hb_002374_470 |
0.1387238278 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 4 |
Hb_012055_080 |
0.1391363866 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 5 |
Hb_005890_020 |
0.1415198489 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 6 |
Hb_004115_010 |
0.1427776125 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 7 |
Hb_011033_010 |
0.1494534207 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 8 |
Hb_000932_110 |
0.1514551132 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 9 |
Hb_002301_120 |
0.1533082357 |
- |
- |
PREDICTED: uncharacterized protein LOC105649581 [Jatropha curcas] |
| 10 |
Hb_000438_160 |
0.1551332332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_059567_010 |
0.1558862294 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 12 |
Hb_003581_230 |
0.1572662316 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 13 |
Hb_000056_030 |
0.157799042 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g14390 isoform X1 [Jatropha curcas] |
| 14 |
Hb_010683_030 |
0.1580086755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000017_200 |
0.1625044512 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |
| 16 |
Hb_000120_930 |
0.1640812418 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_007952_050 |
0.1655194043 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein-like [Jatropha curcas] |
| 18 |
Hb_001511_210 |
0.1663943578 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 19 |
Hb_004143_090 |
0.1671073372 |
- |
- |
PREDICTED: VIN3-like protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_165101_010 |
0.1680610624 |
- |
- |
PREDICTED: putative wall-associated receptor kinase-like 16 [Nelumbo nucifera] |