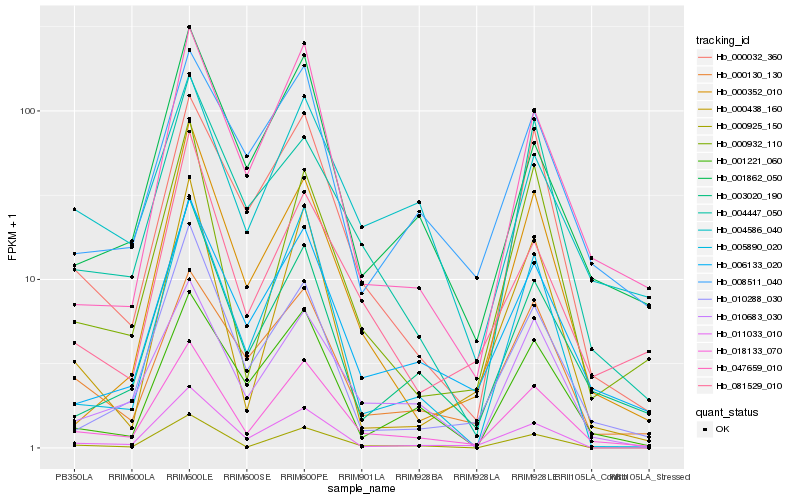
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018133_070 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 2 |
Hb_006133_020 |
0.1133220212 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 3 |
Hb_010683_030 |
0.1238707805 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000032_360 |
0.1274660491 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit S, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000925_150 |
0.1287895235 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 6 |
Hb_003020_190 |
0.1296211559 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 7 |
Hb_047659_010 |
0.1361368143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_011033_010 |
0.1361561244 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 9 |
Hb_000932_110 |
0.1373699032 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 10 |
Hb_000130_130 |
0.138392937 |
- |
- |
hypothetical protein POPTR_0001s40330g [Populus trichocarpa] |
| 11 |
Hb_001862_050 |
0.1427223147 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 12 |
Hb_008511_040 |
0.1442909472 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 13 |
Hb_005890_020 |
0.1447273421 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 14 |
Hb_001221_060 |
0.1448687512 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 15 |
Hb_004586_040 |
0.1490153486 |
transcription factor |
TF Family: MIKC |
K-box region and MADS-box transcription factor family protein [Theobroma cacao] |
| 16 |
Hb_081529_010 |
0.1506191442 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_010288_030 |
0.1521819519 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 18 |
Hb_004447_050 |
0.1530535952 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000438_160 |
0.155560131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000352_010 |
0.1596612798 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |