| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
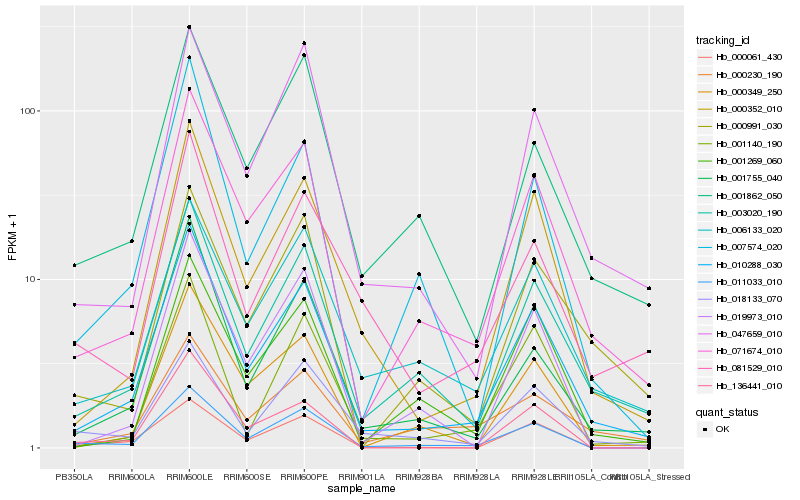
Hb_010288_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 2 |
Hb_071674_010 |
0.0801940403 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 3 |
Hb_003020_190 |
0.0933574858 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 4 |
Hb_000352_010 |
0.0955272419 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 5 |
Hb_011033_010 |
0.1177756663 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 6 |
Hb_001755_040 |
0.12050161 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_007574_020 |
0.1328138302 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 8 |
Hb_006133_020 |
0.1337247893 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 9 |
Hb_000349_250 |
0.1344214346 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 10 |
Hb_047659_010 |
0.1404042545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001269_060 |
0.1443285085 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000061_430 |
0.1473438056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001140_190 |
0.1488120046 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 14 |
Hb_000230_190 |
0.1491766104 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001862_050 |
0.1495195164 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 16 |
Hb_019973_010 |
0.1501761565 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 17 |
Hb_018133_070 |
0.1521819519 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 18 |
Hb_081529_010 |
0.1527243145 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_000991_030 |
0.1532967926 |
- |
- |
PREDICTED: uncharacterized protein LOC105122436 [Populus euphratica] |
| 20 |
Hb_136441_010 |
0.1556193925 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |