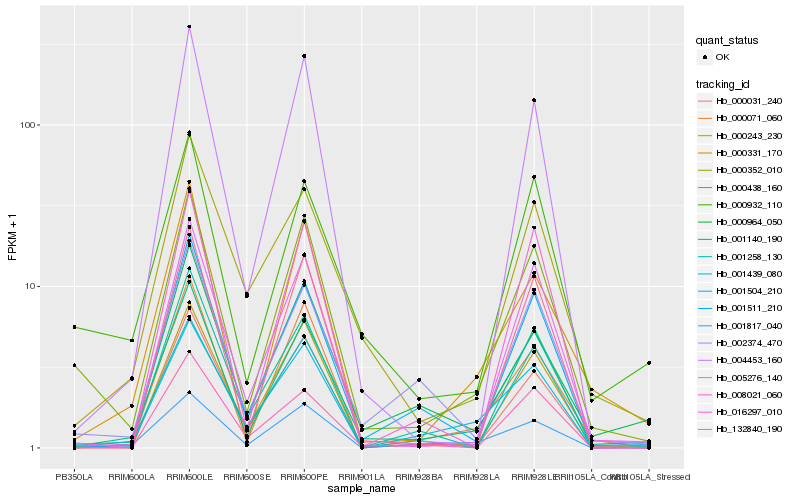
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001140_190 |
0.0 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 2 |
Hb_001511_210 |
0.1033999656 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 3 |
Hb_000438_160 |
0.1176564169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005276_140 |
0.1202761808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000071_060 |
0.1221868479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000331_170 |
0.1227514374 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 7 |
Hb_001817_040 |
0.1247596479 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 8 |
Hb_002374_470 |
0.1275213162 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 9 |
Hb_000031_240 |
0.1304143664 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 10 |
Hb_001258_130 |
0.1308409255 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 11 |
Hb_132840_190 |
0.1310783031 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000352_010 |
0.1315032568 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000932_110 |
0.1327462926 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 14 |
Hb_001439_080 |
0.1333270464 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 15 |
Hb_000964_050 |
0.1338909471 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 16 |
Hb_000243_230 |
0.1357569364 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_004453_160 |
0.1385455874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001504_210 |
0.1396931 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 19 |
Hb_008021_060 |
0.1400044319 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 20 |
Hb_016297_010 |
0.1405592783 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |