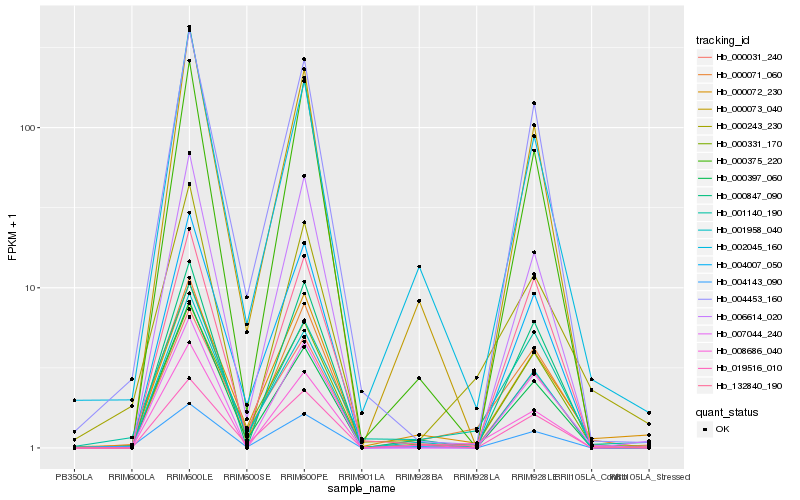
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000071_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000243_230 |
0.1039120992 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_008686_040 |
0.1059743053 |
- |
- |
PREDICTED: uncharacterized protein LOC105632278 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000331_170 |
0.1092897989 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 5 |
Hb_000073_040 |
0.1180195566 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 6 |
Hb_000375_220 |
0.118084509 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 7 |
Hb_000072_230 |
0.1181975453 |
- |
- |
hypothetical protein POPTR_0004s10790g [Populus trichocarpa] |
| 8 |
Hb_007044_240 |
0.1189146427 |
- |
- |
PREDICTED: uncharacterized protein LOC105638704 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004453_160 |
0.119673214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000031_240 |
0.1218084458 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 11 |
Hb_000847_090 |
0.1220130053 |
- |
- |
PREDICTED: uncharacterized protein LOC105644263 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001140_190 |
0.1221868479 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 13 |
Hb_019516_010 |
0.1233588308 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |
| 14 |
Hb_004007_050 |
0.1251817051 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT [Jatropha curcas] |
| 15 |
Hb_001958_040 |
0.1257845345 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 16 |
Hb_002045_160 |
0.1271546153 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 17 |
Hb_132840_190 |
0.1284944221 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_004143_090 |
0.1291627613 |
- |
- |
PREDICTED: VIN3-like protein 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000397_060 |
0.1297478891 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 20 |
Hb_006614_020 |
0.1306898114 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |