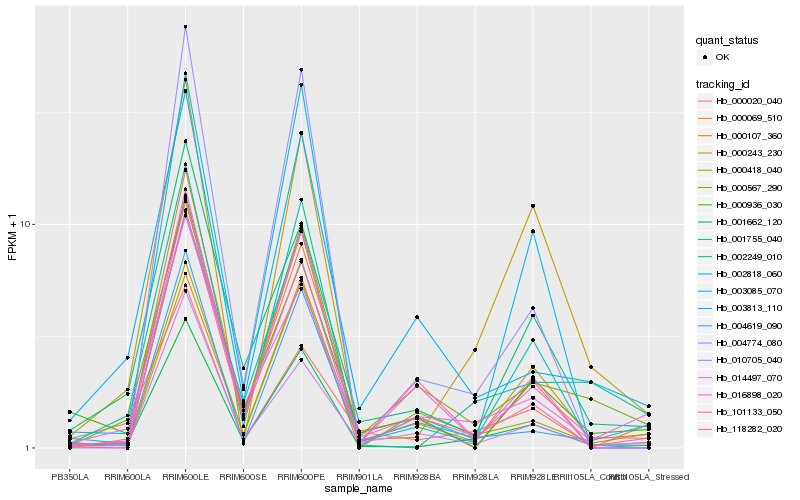
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001662_120 |
0.0 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 2 |
Hb_002249_010 |
0.1122179638 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_118282_020 |
0.144221556 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X3 [Jatropha curcas] |
| 4 |
Hb_001755_040 |
0.1461692567 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_014497_070 |
0.1467013056 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |
| 6 |
Hb_000243_230 |
0.1500739824 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_002818_060 |
0.1504106063 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_000567_290 |
0.1540190897 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 9 |
Hb_000020_040 |
0.1549819562 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_000936_030 |
0.1558715001 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 11 |
Hb_004774_080 |
0.1563429555 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003085_070 |
0.1583319657 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000418_040 |
0.1588994692 |
- |
- |
hypothetical protein RCOM_0377590 [Ricinus communis] |
| 14 |
Hb_004619_090 |
0.1594825671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_010705_040 |
0.1596205611 |
- |
- |
PREDICTED: kinesin-4 [Jatropha curcas] |
| 16 |
Hb_003813_110 |
0.1641935258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_016898_020 |
0.1648853423 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105120899 [Populus euphratica] |
| 18 |
Hb_101133_050 |
0.1651281264 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 19 |
Hb_000069_510 |
0.1673287011 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 20 |
Hb_000107_360 |
0.1691324305 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |