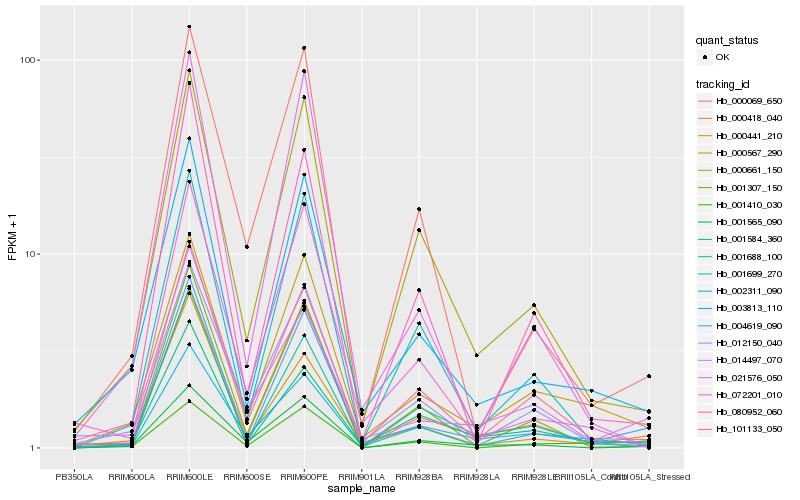
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003813_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000567_290 |
0.0959977692 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 3 |
Hb_000661_150 |
0.1048538291 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 4 |
Hb_004619_090 |
0.1050913942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000441_210 |
0.1117734308 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 6 |
Hb_002311_090 |
0.125546289 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 7 |
Hb_000418_040 |
0.125983813 |
- |
- |
hypothetical protein RCOM_0377590 [Ricinus communis] |
| 8 |
Hb_101133_050 |
0.1261378049 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 9 |
Hb_072201_010 |
0.1265055987 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 10 |
Hb_001584_360 |
0.1269023338 |
- |
- |
PREDICTED: serine/threonine-protein kinase haspin homolog hrk1 [Jatropha curcas] |
| 11 |
Hb_012150_040 |
0.1279172461 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001699_270 |
0.1332748761 |
- |
- |
PREDICTED: uncharacterized protein LOC105628996 [Jatropha curcas] |
| 13 |
Hb_001688_100 |
0.1363313271 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 14 |
Hb_021576_050 |
0.1400041036 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 15 |
Hb_001307_150 |
0.1407505915 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 16 |
Hb_001565_090 |
0.1408747135 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000069_650 |
0.1443983579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_014497_070 |
0.1493788433 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |
| 19 |
Hb_001410_030 |
0.1494139722 |
- |
- |
hypothetical protein JCGZ_20419 [Jatropha curcas] |
| 20 |
Hb_080952_060 |
0.1518429349 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |