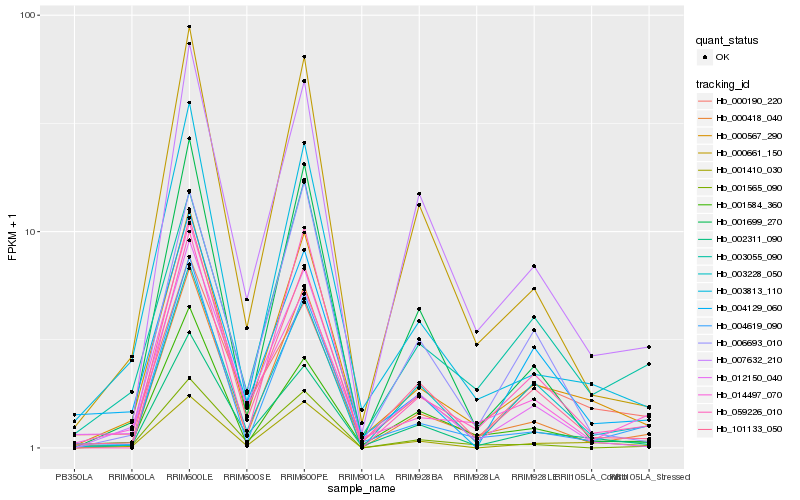
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000567_290 |
0.0 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 2 |
Hb_002311_090 |
0.0944032716 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 3 |
Hb_003813_110 |
0.0959977692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001410_030 |
0.1042594024 |
- |
- |
hypothetical protein JCGZ_20419 [Jatropha curcas] |
| 5 |
Hb_000418_040 |
0.1185139778 |
- |
- |
hypothetical protein RCOM_0377590 [Ricinus communis] |
| 6 |
Hb_101133_050 |
0.1192662852 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 7 |
Hb_000661_150 |
0.1193174477 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 8 |
Hb_012150_040 |
0.1330001349 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007632_210 |
0.1374821132 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |
| 10 |
Hb_004619_090 |
0.1375863056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003055_090 |
0.1431048046 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 12 |
Hb_059226_010 |
0.1441971234 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 13 |
Hb_000190_220 |
0.1449479634 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 14 |
Hb_001584_360 |
0.1454315432 |
- |
- |
PREDICTED: serine/threonine-protein kinase haspin homolog hrk1 [Jatropha curcas] |
| 15 |
Hb_001565_090 |
0.1489533343 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001699_270 |
0.1489760084 |
- |
- |
PREDICTED: uncharacterized protein LOC105628996 [Jatropha curcas] |
| 17 |
Hb_003228_050 |
0.1496133573 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 18 |
Hb_014497_070 |
0.1497904212 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |
| 19 |
Hb_004129_060 |
0.1502554505 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 20 |
Hb_006693_010 |
0.1513266567 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |