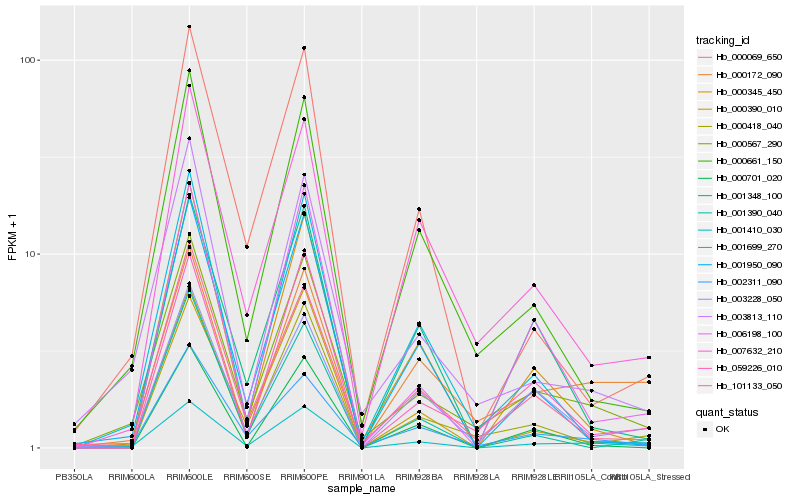
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001410_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_20419 [Jatropha curcas] |
| 2 |
Hb_000567_290 |
0.1042594024 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 3 |
Hb_002311_090 |
0.1304063073 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 4 |
Hb_000418_040 |
0.1433421156 |
- |
- |
hypothetical protein RCOM_0377590 [Ricinus communis] |
| 5 |
Hb_000390_010 |
0.1458690938 |
- |
- |
PREDICTED: protein CDI [Jatropha curcas] |
| 6 |
Hb_003813_110 |
0.1494139722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001950_090 |
0.1499041266 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 8 |
Hb_000345_450 |
0.1510651098 |
- |
- |
PREDICTED: microtubule-binding protein TANGLED [Jatropha curcas] |
| 9 |
Hb_000661_150 |
0.154164618 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 10 |
Hb_059226_010 |
0.1542413451 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 11 |
Hb_000069_650 |
0.154825675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006198_100 |
0.1568722055 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 13 |
Hb_101133_050 |
0.1579916943 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 14 |
Hb_003228_050 |
0.1585677687 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 15 |
Hb_001699_270 |
0.1601550383 |
- |
- |
PREDICTED: uncharacterized protein LOC105628996 [Jatropha curcas] |
| 16 |
Hb_000172_090 |
0.1617300108 |
- |
- |
PREDICTED: uncharacterized protein LOC105650755 [Jatropha curcas] |
| 17 |
Hb_007632_210 |
0.1620090973 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |
| 18 |
Hb_001390_040 |
0.165067733 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 19 |
Hb_000701_020 |
0.166059358 |
- |
- |
PREDICTED: protein ENDOSPERM DEFECTIVE 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001348_100 |
0.1670896181 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |